Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations
- PMID: 19270083
- PMCID: PMC2694482
- DOI: 10.1101/gr.084848.108
Complete genome of the cellulolytic thermophile Acidothermus cellulolyticus 11B provides insights into its ecophysiological and evolutionary adaptations
Abstract
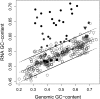
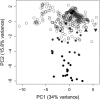
We present here the complete 2.4-Mb genome of the cellulolytic actinobacterial thermophile Acidothermus cellulolyticus 11B. New secreted glycoside hydrolases and carbohydrate esterases were identified in the genome, revealing a diverse biomass-degrading enzyme repertoire far greater than previously characterized and elevating the industrial value of this organism. A sizable fraction of these hydrolytic enzymes break down plant cell walls, and the remaining either degrade components in fungal cell walls or metabolize storage carbohydrates such as glycogen and trehalose, implicating the relative importance of these different carbon sources. Several of the A. cellulolyticus secreted cellulolytic and xylanolytic enzymes are fused to multiple tandemly arranged carbohydrate binding modules (CBM), from families 2 and 3. For the most part, thermophilic patterns in the genome and proteome of A. cellulolyticus were weak, which may be reflective of the recent evolutionary history of A. cellulolyticus since its divergence from its closest phylogenetic neighbor Frankia, a mesophilic plant endosymbiont and soil dweller. However, ribosomal proteins and noncoding RNAs (rRNA and tRNAs) in A. cellulolyticus showed thermophilic traits suggesting the importance of adaptation of cellular translational machinery to environmental temperature. Elevated occurrence of IVYWREL amino acids in A. cellulolyticus orthologs compared to mesophiles and inverse preferences for G and A at the first and third codon positions also point to its ongoing thermoadaptation. Additional interesting features in the genome of this cellulolytic, hot-springs-dwelling prokaryote include a low occurrence of pseudogenes or mobile genetic elements, an unexpected complement of flagellar genes, and the presence of three laterally acquired genomic islands of likely ecophysiological value.
Figures



References
-
- Adney W.S., Tucker M.P., Nieves R.A., Thomas S.R., Himmel M.E. Low molecular weight thermostable β-D-glucosidase from Acidothermus cellulolyticus. Biotechnol. Lett. 1995;17:49–54.
-
- Alloisio N., Marechal J., Heuvel B.V., Normand P., Berry A.M. Characterization of a gene locus containing squalene-hopene cyclase (shc) in Frankia alni ACN14a, and an shc homolog in Acidothermus cellulolyticus. Symbiosis. 2005;39:83–90.
-
- Baker J.O., Adney W.S., Nieves R.A., Thomas S.R., Himmel M.E., Wilson D.B. A new thermostable endoglucanase, Acidothermus cellulolyticus E1. Appl. Biochem. Biotechnol. 1994;45–46:245–256.
-
- Bendtsen J.D., Nielsen H., von Heijne G., Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
