Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae
- PMID: 19270090
- PMCID: PMC2681789
- DOI: 10.1128/JB.01505-08
Influences of capsule on cell shape and chain formation of wild-type and pcsB mutants of serotype 2 Streptococcus pneumoniae
Abstract
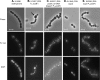
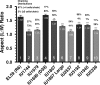
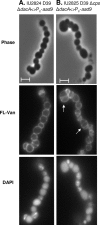
PcsB is a protein of unknown function that plays a critical role in cell division in Streptococcus pneumoniae and other ovococcus species of Streptococcus. We constructed isogenic sets of mutants expressing different amounts of PcsB in laboratory strain R6 and virulent serotype 2 strain D39 to evaluate its cellular roles. Insertion mutagenesis in parent and pcsB(+) merodiploid strains indicated that pcsB is essential in serotype 2 S. pneumoniae. Quantitative Western blotting of wild-type and epitope-tagged PcsB showed that all PcsB was processed into cell-associated and secreted forms of the same molecular mass and that cell-associated PcsB was moderately abundant and present at approximately 4,900 monomers per cell. Controlled expression and complementation experiments indicated that there was a causative relationship between the severity of defects in cell division and decreasing PcsB amount. These experiments also showed that perturbations of expression of the upstream mreCD genes did not contribute to the cell division defects of pcsB mutants and that mreCD could be deleted. Unexpectedly, capsule influenced the cell shape and chain formation phenotypes of the wild-type D39 strain and mutants underexpressing PcsB or deleted for other genes involved in peptidoglycan biosynthesis, such as dacA. Underexpression of PcsB did not result in changes in the amounts or composition of lactoyl-peptides, which were markedly different in the R6 and D39 strains, and there was no correlation between decreased PcsB amount and sensitivity to penicillin. Finally, microarray analyses indicated that underexpression of PcsB may generate a signal that increases expression of the VicRK regulon, which includes pcsB.
Figures




References
-
- Arbeloa, A., J. E. Hugonnet, A. C. Sentilhes, N. Josseaume, L. Dubost, C. Monsempes, D. Blanot, J. P. Brouard, and M. Arthur. 2004. Synthesis of mosaic peptidoglycan cross-bridges by hybrid peptidoglycan assembly pathways in gram-positive bacteria. J. Biol. Chem. 27941546-41556. - PubMed
-
- Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28234-237. - PubMed
-
- Battig, P., and K. Muhlemann. 2007. Capsule genes of Streptococcus pneumoniae influence growth in vitro. FEMS Immunol. Med. Microbiol. 50324-329. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

