Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome
- PMID: 19270159
- PMCID: PMC2658520
- DOI: 10.1101/gad.511809
Chromatin state marks cell-type- and gender-specific replication of the Drosophila genome
Abstract
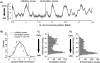
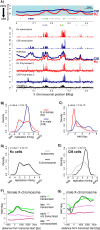
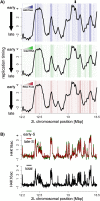
Duplication of eukaryotic genomes during S phase is coordinated in space and time. In order to identify zones of initiation and cell-type- as well as gender-specific plasticity of DNA replication, we profiled replication timing, histone acetylation, and transcription throughout the Drosophila genome. We observed two waves of replication initiation with many distinct zones firing in early-S phase and multiple, less defined peaks at the end of S phase, suggesting that initiation becomes more promiscuous in late-S phase. A comparison of different cell types revealed widespread plasticity of replication timing on autosomes. Most occur in large regions, but only half coincide with local differences in transcription. In contrast to confined autosomal differences, a global shift in replication timing occurs throughout the single male X chromosome. Unlike in females, the dosage-compensated X chromosome replicates almost exclusively early. This difference occurs at sites that are not transcriptionally hyperactivated, but show increased acetylation of Lys 16 of histone H4 (H4K16ac). This suggests a transcription-independent, yet chromosome-wide process related to chromatin. Importantly, H4K16ac is also enriched at initiation zones as well as early replicating regions on autosomes during S phase. Together, our study reveals novel organizational principles of DNA replication of the Drosophila genome and suggests that H4K16ac is more closely correlated with replication timing than is transcription.
Figures


References
-
- Aggarwal B.D., Calvi B.R. Chromatin regulates origin activity in Drosophila follicle cells. Nature. 2004;430:372–376. - PubMed
-
- Aladjem M.I. Replication in context: Dynamic regulation of DNA replication patterns in metazoans. Nat. Rev. Genet. 2007;8:588–600. - PubMed
-
- Aladjem M.I., Rodewald L.W., Kolman J.L., Wahl G.M. Genetic dissection of a mammalian replicator in the human β-globin locus. Science. 1998;281:1005–1009. - PubMed
-
- Anglana M., Apiou F., Bensimon A., Debatisse M. Dynamics of DNA replication in mammalian somatic cells: Nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003;114:385–394. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
