Inverse regulation of plasticity-related immediate early genes by calcineurin in hippocampal neurons
- PMID: 19270309
- PMCID: PMC2673322
- DOI: 10.1074/jbc.M901121200
Inverse regulation of plasticity-related immediate early genes by calcineurin in hippocampal neurons
Abstract
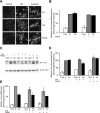
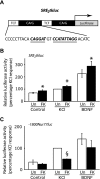
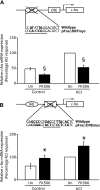
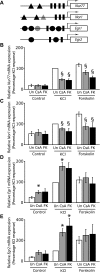
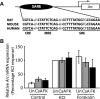
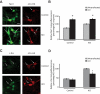
In the mammalian hippocampus, changes in the expression of immediate early genes (IEGs) is thought to contribute to long term plastic changes in neurons brought about by learning tasks and high frequency stimulation of synapses. The phosphatase calcineurin has emerged as an important negative regulator of hippocampus-dependent learning and long term potentiation. Here we investigated the possibility that the constraining action of calcineurin on hippocampal plasticity is mediated in part by regulation of gene expression through negative control of transcription factors, such as cAMP-response element (CRE)-binding protein (CREB). We assessed the effect of calcineurin inhibitors on CREB activation by neuronal activity and show that calcineurin activity is in fact required for CREB-mediated gene expression. However, inhibition of calcineurin had disparate effects on the transcriptional induction of CREB-dependent IEGs. We find that the IEG c-fos is unaffected by suppression of calcineurin activity, the plasticity-related genes Egr1/Zif268 and Egr2/Krox-20 are up-regulated, and genes encoding the orphan nuclear hormone receptors Nor1 and Nur77 are down-regulated. We further show that the up-regulation of particular IEGs is probably due to the presence of serum response elements (SREs) in their promoters, because SRE-mediated gene expression is enhanced by calcineurin blockers. Moreover, expression of the c-fos gene, which is unaffected by calcineurin inhibitors, could be down-regulated by mutating the SRE. Conversely, SRE-mediated c-fos induction in the absence of a functional CRE was enhanced by calcineurin inhibitors. Our experiments thus implicate calcineurin as a negative regulator of SRE-dependent neuronal genes.
Figures
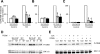
Similar articles
-
Subfield-specific immediate early gene expression associated with hippocampal long-term potentiation in vivo.Eur J Neurosci. 2001 Mar;13(5):968-76. doi: 10.1046/j.0953-816x.2001.01467.x. Eur J Neurosci. 2001. PMID: 11264669
-
Neuromodulatory effect of Gαs- or Gαq-coupled G-protein-coupled receptor on NMDA receptor selectively activates the NMDA receptor/Ca2+/calcineurin/cAMP response element-binding protein-regulated transcriptional coactivator 1 pathway to effectively induce brain-derived neurotrophic factor expression in neurons.J Neurosci. 2015 Apr 8;35(14):5606-24. doi: 10.1523/JNEUROSCI.3650-14.2015. J Neurosci. 2015. PMID: 25855176 Free PMC article.
-
Extracellular signal-regulated kinase (ERK) controls immediate early gene induction on corticostriatal stimulation.J Neurosci. 1998 Nov 1;18(21):8814-25. doi: 10.1523/JNEUROSCI.18-21-08814.1998. J Neurosci. 1998. PMID: 9786988 Free PMC article.
-
Shaping synaptic plasticity: the role of activity-mediated epigenetic regulation on gene transcription.Int J Dev Neurosci. 2013 Oct;31(6):359-69. doi: 10.1016/j.ijdevneu.2013.04.003. Epub 2013 May 9. Int J Dev Neurosci. 2013. PMID: 23665156 Review.
-
A role for immediate-early transcription factors in learning and memory.Behav Genet. 1996 May;26(3):293-9. doi: 10.1007/BF02359385. Behav Genet. 1996. PMID: 8754252 Review.
Cited by
-
Critical Protein-Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription.Molecules. 2021 Oct 11;26(20):6125. doi: 10.3390/molecules26206125. Molecules. 2021. PMID: 34684708 Free PMC article. Review.
-
Nr4a1-eGFP is a marker of striosome-matrix architecture, development and activity in the extended striatum.PLoS One. 2011 Jan 28;6(1):e16619. doi: 10.1371/journal.pone.0016619. PLoS One. 2011. PMID: 21305052 Free PMC article.
-
Molecular mechanism of nur77 gene expression and downstream target genes in the early stage of forskolin-induced differentiation in PC12 cells.Sci Rep. 2020 Apr 14;10(1):6325. doi: 10.1038/s41598-020-62968-y. Sci Rep. 2020. PMID: 32286359 Free PMC article.
-
Calcineurin inhibition may prevent Alzheimer disease in people with Down syndrome.Alzheimers Dement. 2025 Mar;21(3):e70034. doi: 10.1002/alz.70034. Alzheimers Dement. 2025. PMID: 40042516 Free PMC article. Review.
-
The cytochrome c gene proximal enhancer drives activity-dependent reporter gene expression in hippocampal neurons.Front Mol Neurosci. 2012 Mar 7;5:31. doi: 10.3389/fnmol.2012.00031. eCollection 2012. Front Mol Neurosci. 2012. PMID: 22408605 Free PMC article.
References
-
- Lanahan, A., and Worley, P. (1998) Neurobiol. Learn. Mem. 70 37-43 - PubMed
-
- Mansuy, I. M. (2003) Biochem. Biophys. Res. Commun. 311 1195-1208 - PubMed
-
- Zeng, H., Chattarji, S., Barbarosie, M., Rondi-Reig, L., Philpot, B. D., Miyakawa, T., Bear, M. F., and Tonegawa, S. (2001) Cell 107 617-629 - PubMed
-
- Winder, D. G., and Sweatt, J. D. (2001) Nat. Rev. Neurosci. 2 461-474 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

