CBP/p300-mediated acetylation of histone H3 on lysine 56
- PMID: 19270680
- PMCID: PMC2756583
- DOI: 10.1038/nature07861
CBP/p300-mediated acetylation of histone H3 on lysine 56
Erratum in
- Nature. 2009 Aug 27;460(7259):1164
Abstract
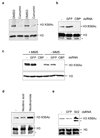
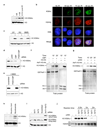
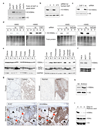
Acetylation within the globular core domain of histone H3 on lysine 56 (H3K56) has recently been shown to have a critical role in packaging DNA into chromatin following DNA replication and repair in budding yeast. However, the function or occurrence of this specific histone mark has not been studied in multicellular eukaryotes, mainly because the Rtt109 enzyme that is known to mediate acetylation of H3K56 (H3K56ac) is fungal-specific. Here we demonstrate that the histone acetyl transferase CBP (also known as Nejire) in flies and CBP and p300 (Ep300) in humans acetylate H3K56, whereas Drosophila Sir2 and human SIRT1 and SIRT2 deacetylate H3K56ac. The histone chaperones ASF1A in humans and Asf1 in Drosophila are required for acetylation of H3K56 in vivo, whereas the histone chaperone CAF-1 (chromatin assembly factor 1) in humans and Caf1 in Drosophila are required for the incorporation of histones bearing this mark into chromatin. We show that, in response to DNA damage, histones bearing acetylated K56 are assembled into chromatin in Drosophila and human cells, forming foci that colocalize with sites of DNA repair. Furthermore, acetylation of H3K56 is increased in multiple types of cancer, correlating with increased levels of ASF1A in these tumours. Our identification of multiple proteins regulating the levels of H3K56 acetylation in metazoans will allow future studies of this critical and unique histone modification that couples chromatin assembly to DNA synthesis, cell proliferation and cancer.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

