Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS
- PMID: 19271777
- PMCID: PMC2707821
- DOI: 10.1021/bi8019614
Structure of the redox sensor domain of Methylococcus capsulatus (Bath) MmoS
Abstract
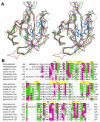
MmoS from Methylococcus capsulatus (Bath) is the multidomain sensor protein of a two-component signaling system proposed to play a role in the copper-mediated regulation of soluble methane monooxygenase (sMMO). MmoS binds an FAD cofactor within its N-terminal tandem Per-Arnt-Sim (PAS) domains, suggesting that it functions as a redox sensor. The crystal structure of the MmoS tandem PAS domains, designated PAS-A and PAS-B, has been determined to 2.34 A resolution. Both domains adopt the typical PAS domain alpha/beta topology and are structurally similar. The two domains are linked by a long alpha helix and do not interact with one another. The FAD cofactor is housed solely within PAS-A and is stabilized by an extended hydrogen bonding network. The overall fold of PAS-A is similar to those of other flavin-containing PAS domains, but homodimeric interactions in other structures are not observed in the MmoS sensor, which crystallized as a monomer. The structure both provides new insight into the architecture of tandem PAS domains and suggests specific residues that may play a role in MmoS FAD redox chemistry and subsequent signal transduction.
Figures



Similar articles
-
Biochemical characterization of MmoS, a sensor protein involved in copper-dependent regulation of soluble methane monooxygenase.Biochemistry. 2006 Aug 29;45(34):10191-8. doi: 10.1021/bi060693h. Biochemistry. 2006. PMID: 16922494
-
Expression and characterization of ferredoxin and flavin adenine dinucleotide binding domains of the reductase component of soluble methane monooxygenase from Methylococcus capsulatus (Bath).Biochemistry. 2002 Dec 31;41(52):15780-94. doi: 10.1021/bi026757f. Biochemistry. 2002. PMID: 12501207
-
NMR structure of the flavin domain from soluble methane monooxygenase reductase from Methylococcus capsulatus (Bath).Biochemistry. 2004 Sep 28;43(38):11983-91. doi: 10.1021/bi049066n. Biochemistry. 2004. PMID: 15379538
-
Enzymatic oxidation of methane.Biochemistry. 2015 Apr 14;54(14):2283-94. doi: 10.1021/acs.biochem.5b00198. Epub 2015 Apr 1. Biochemistry. 2015. PMID: 25806595 Free PMC article. Review.
-
Structural and mechanistic insights into methane oxidation by particulate methane monooxygenase.Acc Chem Res. 2007 Jul;40(7):573-80. doi: 10.1021/ar700004s. Epub 2007 Apr 20. Acc Chem Res. 2007. PMID: 17444606 Review.
Cited by
-
Flavin redox switching of protein functions.Antioxid Redox Signal. 2011 Mar 15;14(6):1079-91. doi: 10.1089/ars.2010.3417. Epub 2010 Oct 28. Antioxid Redox Signal. 2011. PMID: 21028987 Free PMC article. Review.
-
Origin and functional diversification of PAS domain, a ubiquitous intracellular sensor.Sci Adv. 2023 Sep;9(35):eadi4517. doi: 10.1126/sciadv.adi4517. Epub 2023 Aug 30. Sci Adv. 2023. PMID: 37647406 Free PMC article.
-
Diversity in Sensing and Signaling of Bacterial Sensor Histidine Kinases.Biomolecules. 2021 Oct 15;11(10):1524. doi: 10.3390/biom11101524. Biomolecules. 2021. PMID: 34680156 Free PMC article. Review.
-
Methane monooxygenases: central enzymes in methanotrophy with promising biotechnological applications.World J Microbiol Biotechnol. 2021 Mar 25;37(4):72. doi: 10.1007/s11274-021-03038-x. World J Microbiol Biotechnol. 2021. PMID: 33765207 Free PMC article. Review.
-
A PilZ-Containing Chemotaxis Receptor Mediates Oxygen and Wheat Root Sensing in Azospirillum brasilense.Front Microbiol. 2019 Mar 1;10:312. doi: 10.3389/fmicb.2019.00312. eCollection 2019. Front Microbiol. 2019. PMID: 30881352 Free PMC article.
References
-
- Hefti M, Françoijs K-J, de Vries LC, Dixon R, Vervoort J. The PAS fold: a redefinition of the PAS domain based upon structural prediction. Eur. J. Biochem. 2004;271:1198–1208. - PubMed
-
- Ponting CP, Aravind L. PAS: a multifunctional domain family comes to light. Current Biol. 1997;7:R674–R677. - PubMed
-
- Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Ann. Rev. Biochem. 2000;69:183–215. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

