Suppression of HIV-1 replication by microRNA effectors
- PMID: 19272132
- PMCID: PMC2657893
- DOI: 10.1186/1742-4690-6-26
Suppression of HIV-1 replication by microRNA effectors
Abstract
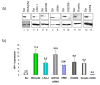
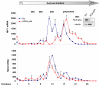
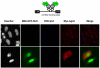
The rate of HIV-1 gene expression is a key step that determines the kinetics of virus spread and AIDS progression. Viral entry and gene expression were described to be the key determinants for cell permissiveness to HIV. Recent reports highlighted the involvement of miRNA in regulating HIV-1 replication post-transcriptionally. In this study we explored the role of cellular factors required for miRNA-mediated mRNA translational inhibition in regulating HIV-1 gene expression. Here we show that HIV-1 mRNAs associate and co-localize with components of the RNA Induced Silencing Complex (RISC), and we characterize some of the proteins required for miRNA-mediated silencing (miRNA effectors). RCK/p54, GW182, LSm-1 and XRN1 negatively regulate HIV-1 gene expression by preventing viral mRNA association with polysomes. Interestingly, knockdown of RCK/p54 or DGCR8 resulted in virus reactivation in PBMCs isolated from HIV infected patients treated with suppressive HAART.
Figures

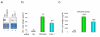

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

