The evolution of vertebrate opioid receptors
- PMID: 19273128
- PMCID: PMC3070387
- DOI: 10.2741/3306
The evolution of vertebrate opioid receptors
Abstract
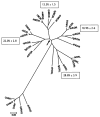
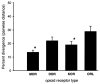
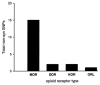
The proteins that mediate the analgesic and other effects of opioid drugs and endogenous opioid peptides are known as opioid receptors. Opioid receptors consist of a family of four closely-related proteins belonging to the large superfamily of G-protein coupled receptors. The three types of opioid receptors shown unequivocally to mediate analgesia in animal models are the mu (MOR), delta (DOR), and kappa (KOR) opioid receptor proteins. The role of the fourth member of the opioid receptor family, the nociceptin or orphanin FQ receptor (ORL), is not as clear as hyperalgesia, analgesia, and no effect was reported after administration of ORL agonists. There are now cDNA sequences for all four types of opioid receptors that are expressed in the brain of six species from three different classes of vertebrates. This review presents a comparative analysis of vertebrate opioid receptors using bioinformatics and data from recent human genome studies. Results indicate that opioid receptors arose by gene duplication, that there is a vector of opioid receptor divergence, and that MOR shows evidence of rapid evolution.
Figures



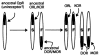
References
-
- Attwood TK. A compendium of specific motifs for diagnosing GPCR subtypes. Trends Pharmacol Sci. 2001;22:162–165. - PubMed
-
- Perez DM. The evolutionarily triumphant g-protein-coupled receptor. Mol Pharmacol. 2003;63:1202–1205. - PubMed
-
- Perez DM. From plants to man: the GPCR “tree of life”. Mol Pharmacol. 2005;67:1383–1384. - PubMed
-
- Fredriksson R, Lagerstrom MC, Lundin LG, Schioth HB. The G-protein-coupled receptors in the human genome form five main families: Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

