RNA-protein interactions in hepadnavirus reverse transcription
- PMID: 19273150
- PMCID: PMC3611959
- DOI: 10.2741/3328
RNA-protein interactions in hepadnavirus reverse transcription
Abstract
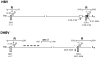
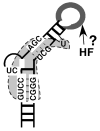
The small DNA genome of hepadnaviruses is replicated by reverse transcription via an RNA intermediate. This RNA "pregenome" contains important signals that control critical steps of viral replication, including RNA packaging, initiation of reverse transcription, and elongation of minus strand DNA, through specific interactions with the viral reverse transcriptase, the capsid protein, and host factors. In particular, the interaction between the viral reverse transcriptase and RNA pregenome requires a host chaperone complex composed of the heat shock protein 90 and its cochaperones.
Figures
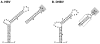
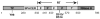
Similar articles
-
Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer.J Virol. 1994 Jun;68(6):3536-43. doi: 10.1128/JVI.68.6.3536-3543.1994. J Virol. 1994. PMID: 8189492 Free PMC article.
-
Identification of a signal necessary for initiation of reverse transcription of the hepadnavirus genome.J Virol. 1991 Oct;65(10):5190-5. doi: 10.1128/JVI.65.10.5190-5195.1991. J Virol. 1991. PMID: 1895379 Free PMC article.
-
Why are hepadnaviruses DNA and not RNA viruses?Trends Microbiol. 1997 Nov;5(11):447-50. doi: 10.1016/s0966-842x(97)01141-4. Trends Microbiol. 1997. PMID: 9402701 Review.
-
Conservation of the HBV RNA element epsilon in nackednaviruses reveals ancient origin of protein-primed reverse transcription.Proc Natl Acad Sci U S A. 2021 Mar 30;118(13):e2022373118. doi: 10.1073/pnas.2022373118. Proc Natl Acad Sci U S A. 2021. PMID: 33753499 Free PMC article.
-
How do viral reverse transcriptases recognize their RNA genome?FEBS Lett. 1991 Aug 5;287(1-2):1-4. doi: 10.1016/0014-5793(91)80002-k. FEBS Lett. 1991. PMID: 1715279 Review.
Cited by
-
Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein.PLoS Pathog. 2013;9(7):e1003494. doi: 10.1371/journal.ppat.1003494. Epub 2013 Jul 11. PLoS Pathog. 2013. PMID: 23853601 Free PMC article.
-
Drastic reduction in the production of subviral particles does not impair hepatitis B virus virion secretion.J Virol. 2009 Nov;83(21):11152-65. doi: 10.1128/JVI.00905-09. Epub 2009 Aug 12. J Virol. 2009. PMID: 19706705 Free PMC article.
-
In vitro epsilon RNA-dependent protein priming activity of human hepatitis B virus polymerase.J Virol. 2012 May;86(9):5134-50. doi: 10.1128/JVI.07137-11. Epub 2012 Feb 29. J Virol. 2012. PMID: 22379076 Free PMC article.
-
Protein-primed terminal transferase activity of hepatitis B virus polymerase.J Virol. 2013 Mar;87(5):2563-76. doi: 10.1128/JVI.02786-12. Epub 2012 Dec 19. J Virol. 2013. PMID: 23255788 Free PMC article.
-
RNA-Binding Motif Protein 24 (RBM24) Is Involved in Pregenomic RNA Packaging by Mediating Interaction between Hepatitis B Virus Polymerase and the Epsilon Element.J Virol. 2019 Mar 5;93(6):e02161-18. doi: 10.1128/JVI.02161-18. Print 2019 Mar 15. J Virol. 2019. PMID: 30626666 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

