Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes
- PMID: 19273586
- PMCID: PMC2682052
- DOI: 10.1128/MCB.01682-08
Mutational analysis of the Sir3 BAH domain reveals multiple points of interaction with nucleosomes
Abstract
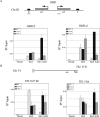
Sir3, a component of the transcriptional silencing complex in the yeast Saccharomyces cerevisiae, has an N-terminal BAH domain that is crucial for the protein's silencing function. Previous work has shown that the N-terminal alanine residue of Sir3 (Ala2) and its acetylation play an important role in silencing. Here we show that the silencing defects of Sir3 Ala2 mutants can be suppressed by mutations in histones H3 and H4, specifically, by H3 D77N and H4 H75Y mutations. Additionally, a mutational analysis demonstrates that three separate regions of the Sir3 BAH domain are important for its role in silencing. Many of these BAH mutations also can be suppressed by the H3 D77N and H4 H75Y mutations. In agreement with the results of others, in vitro experiments show that the Sir3 BAH domain can interact with partially purified nucleosomes. The silencing-defective BAH mutants are defective for this interaction. These results, together with the previously characterized interaction between the C-terminal region of Sir3 and the histone H3/H4 tails, suggest that Sir3 utilizes multiple domains to interact with nucleosomes.
Figures





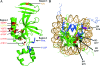
Similar articles
-
Structure and function of the Saccharomyces cerevisiae Sir3 BAH domain.Mol Cell Biol. 2006 Apr;26(8):3256-65. doi: 10.1128/MCB.26.8.3256-3265.2006. Mol Cell Biol. 2006. PMID: 16581798 Free PMC article.
-
Compensatory interactions between Sir3p and the nucleosomal LRS surface imply their direct interaction.PLoS Genet. 2008 Dec;4(12):e1000301. doi: 10.1371/journal.pgen.1000301. Epub 2008 Dec 12. PLoS Genet. 2008. PMID: 19079580 Free PMC article.
-
Sir3-nucleosome interactions in spreading of silent chromatin in Saccharomyces cerevisiae.Mol Cell Biol. 2008 Nov;28(22):6903-18. doi: 10.1128/MCB.01210-08. Epub 2008 Sep 15. Mol Cell Biol. 2008. PMID: 18794362 Free PMC article.
-
Heterochromatin protein Sir3 induces contacts between the amino terminus of histone H4 and nucleosomal DNA.Proc Natl Acad Sci U S A. 2013 May 21;110(21):8495-500. doi: 10.1073/pnas.1300126110. Epub 2013 May 6. Proc Natl Acad Sci U S A. 2013. PMID: 23650358 Free PMC article.
-
Epigenetics in Saccharomyces cerevisiae.Cold Spring Harb Perspect Biol. 2013 Jul 1;5(7):a017491. doi: 10.1101/cshperspect.a017491. Cold Spring Harb Perspect Biol. 2013. PMID: 23818500 Free PMC article. Review.
Cited by
-
Sir3 Polymorphisms in Candida glabrata clinical isolates.Mycopathologia. 2013 Apr;175(3-4):207-19. doi: 10.1007/s11046-013-9627-2. Epub 2013 Feb 8. Mycopathologia. 2013. PMID: 23392823
-
Reinventing heterochromatin in budding yeasts: Sir2 and the origin recognition complex take center stage.Eukaryot Cell. 2011 Sep;10(9):1183-92. doi: 10.1128/EC.05123-11. Epub 2011 Jul 15. Eukaryot Cell. 2011. PMID: 21764908 Free PMC article. Review.
-
The biological functions of Naa10 - From amino-terminal acetylation to human disease.Gene. 2015 Aug 10;567(2):103-31. doi: 10.1016/j.gene.2015.04.085. Epub 2015 May 16. Gene. 2015. PMID: 25987439 Free PMC article. Review.
-
Symmetry, asymmetry, and kinetics of silencing establishment in Saccharomyces cerevisiae revealed by single-cell optical assays.Proc Natl Acad Sci U S A. 2011 Jan 25;108(4):1209-16. doi: 10.1073/pnas.1018742108. Epub 2011 Jan 24. Proc Natl Acad Sci U S A. 2011. PMID: 21262833 Free PMC article.
-
Regulation of telomere silencing by the core histones-autophagy-Sir2 axis.Life Sci Alliance. 2022 Dec 30;6(3):e202201614. doi: 10.26508/lsa.202201614. Print 2023 Mar. Life Sci Alliance. 2022. PMID: 36585257 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. Kingston, D. Moore, J. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
-
- Burke, D., D. Dawson, T. Stearns, and Cold Spring Harbor Laboratory. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual, 2000 ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
-
- Callebaut, I., J. C. Courvalin, and J. P. Mornon. 1999. The BAH (bromo-adjacent homology) domain: a link between DNA methylation, replication and transcriptional regulation. FEBS Lett. 446189-193. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
