Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response
- PMID: 19273602
- PMCID: PMC2682049
- DOI: 10.1128/MCB.01639-08
Acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 during the antioxidant response
Abstract
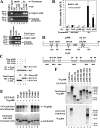
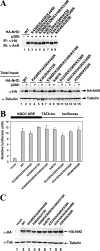
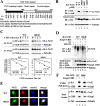
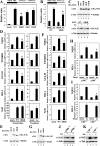
To maintain intracellular redox homeostasis, genes encoding many antioxidants and detoxification enzymes are transcriptionally upregulated upon deleterious oxidative stress through the cis antioxidant responsive elements (AREs) in their promoter regions. Nrf2 is the critical transcription factor responsible for ARE-dependent transcription. We and others have previously demonstrated that Nrf2 is targeted for ubiquitin-mediated degradation by Keap1 in a redox-sensitive manner through modifications of distinct cysteine residues of Keap1. Here, we report that p300/CBP directly acetylates Nrf2 in response to arsenite-induced stress. We have identified multiple acetylated lysine residues within the Nrf2 Neh1 DNA-binding domain. Combined lysine-to-arginine mutations on the acetylation sites, with no effects on Nrf2 protein stability, compromised the DNA-binding activity of Nrf2 in a promoter-specific manner. These findings demonstrated that acetylation of Nrf2 by p300/CBP augments promoter-specific DNA binding of Nrf2 and established acetylation as a novel regulatory mechanism that functions in concert with Keap1-mediated ubiquitination in modulating the Nrf2-dependent antioxidant response.
Figures



References
-
- Alam, J., and J. L. Cook. 2007. How many transcription factors does it take to turn on the heme oxygenase-1 gene? Am. J. Respir. Cell Mol. Biol. 36166-174. - PubMed
-
- Alam, J., D. Stewart, C. Touchard, S. Boinapally, A. M. Choi, and J. L. Cook. 1999. Nrf2, a Cap′n'Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J. Biol. Chem. 27426071-26078. - PubMed
-
- Ben-Shahar, T. R., S. Heeger, C. Lehane, P. East, H. Flynn, M. Skehel, and F. Uhlmann. 2008. EcoI-dependent cohesin acetylation during establishment of sister chromatid cohesion. Science 321563-566. - PubMed
-
- Brunet, A., L. B. Sweeney, J. F. Sturgill, K. F. Chua, P. L. Greer, Y. Lin, H. Tran, S. E. Ross, R. Mostoslavsky, H. Y. Cohen, L. S. Hu, H. L. Cheng, M. P. Jedrychowski, S. P. Gygi, D. A. Sinclair, F. W. Alt, and M. E. Greenberg. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 3032011-2015. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
