Cholesterol metabolism: the main pathway acting downstream of cytochrome P450 oxidoreductase in skeletal development of the limb
- PMID: 19273610
- PMCID: PMC2682028
- DOI: 10.1128/MCB.01638-08
Cholesterol metabolism: the main pathway acting downstream of cytochrome P450 oxidoreductase in skeletal development of the limb
Abstract
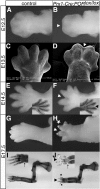
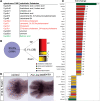
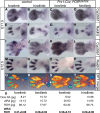
Cytochrome P450 oxidoreductase (POR) is the obligate electron donor for all microsomal cytochrome P450 enzymes, which catalyze the metabolism of a wide spectrum of xenobiotic and endobiotic compounds. Point mutations in POR have been found recently in patients with Antley-Bixler-like syndrome, which includes limb skeletal defects. In order to study P450 function during limb and skeletal development, we deleted POR specifically in mouse limb bud mesenchyme. Forelimbs and hind limbs in conditional knockout (CKO) mice were short with thin skeletal elements and fused joints. POR deletion occurred earlier in forelimbs than in hind limbs, leading additionally to soft tissue syndactyly and loss of wrist elements and phalanges due to changes in growth, cell death, and skeletal segmentation. Transcriptional analysis of E12.5 mouse forelimb buds demonstrated the expression of P450s involved in retinoic acid, cholesterol, and arachidonic acid metabolism. Biochemical analysis of CKO limbs confirmed retinoic acid excess. In CKO limbs, expression of genes throughout the whole cholesterol biosynthetic pathway was upregulated, and cholesterol deficiency can explain most aspects of the phenotype. Thus, cellular POR-dependent cholesterol synthesis is essential during limb and skeletal development. Modulation of P450 activity could contribute to susceptibility of the embryo and developing organs to teratogenesis.
Figures

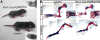




References
-
- Bijlsma, M. F., K. S. Borensztajn, H. Roelink, M. P. Peppelenbosch, and C. A. Spek. 2007. Sonic hedgehog induces transcription-independent cytoskeletal rearrangement and migration regulated by arachidonate metabolites. Cell Signal. 192596-2604. - PubMed
-
- Black, S. D., and M. J. Coon. 1987. P-450 cytochromes: structure and function. Adv. Enzymol. Relat. Areas Mol. Biol. 6035-87. - PubMed
-
- Blomhoff, R., and H. K. Blomhoff. 2006. Overview of retinoid metabolism and function. J. Neurobiol. 66606-630. - PubMed
-
- Bogatcheva, N. V., M. G. Sergeeva, S. M. Dudek, and A. D. Verin. 2005. Arachidonic acid cascade in endothelial pathobiology. Microvasc. Res. 69107-127. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
