ER membrane-bending proteins are necessary for de novo nuclear pore formation
- PMID: 19273614
- PMCID: PMC2686408
- DOI: 10.1083/jcb.200806174
ER membrane-bending proteins are necessary for de novo nuclear pore formation
Abstract
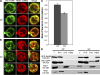
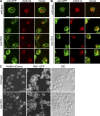
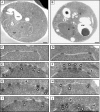
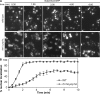
Nucleocytoplasmic transport occurs exclusively through nuclear pore complexes (NPCs) embedded in pores formed by inner and outer nuclear membrane fusion. The mechanism for de novo pore and NPC biogenesis remains unclear. Reticulons (RTNs) and Yop1/DP1 are conserved membrane protein families required to form and maintain the tubular endoplasmic reticulum (ER) and the postmitotic nuclear envelope. In this study, we report that members of the RTN and Yop1/DP1 families are required for nuclear pore formation. Analysis of Saccharomyces cerevisiae prp20-G282S and nup133 Delta NPC assembly mutants revealed perturbations in Rtn1-green fluorescent protein (GFP) and Yop1-GFP ER distribution and colocalization to NPC clusters. Combined deletion of RTN1 and YOP1 resulted in NPC clustering, nuclear import defects, and synthetic lethality with the additional absence of Pom34, Pom152, and Nup84 subcomplex members. We tested for a direct role in NPC biogenesis using Xenopus laevis in vitro assays and found that anti-Rtn4a antibodies specifically inhibited de novo nuclear pore formation. We hypothesize that these ER membrane-bending proteins mediate early NPC assembly steps.
Figures



References
-
- Abramoff M., Magelhaes P., Ram S. 2004. Image processing with ImageJ.Biophotonics International. 11:36–42
-
- Aitchison J.D., Rout M.P., Marelli M., Blobel G., Wozniak R.W. 1995. Two novel related yeast nucleoporins Nup170p and Nup157p: complementation with the vertebrate homologue Nup155p and functional interactions with the yeast nuclear pore-membrane protein Pom152p.J. Cell Biol. 131:1133–1148 - PMC - PubMed
-
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007a. Determining the architectures of macromolecular assemblies.Nature. 450:683–694 - PubMed
-
- Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T., et al. 2007b. The molecular architecture of the nuclear pore complex.Nature. 450:695–701 - PubMed
-
- Anderson D.J., Hetzer M.W. 2007. Nuclear envelope formation by chromatin-mediated reorganization of the endoplasmic reticulum.Nat. Cell Biol. 9:1160–1166 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM057438/GM/NIGMS NIH HHS/United States
- P60 DK020593/DK/NIDDK NIH HHS/United States
- DK58404/DK/NIDDK NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- P30 HD015052/HD/NICHD NIH HHS/United States
- EY08126/EY/NEI NIH HHS/United States
- R01 GM57438/GM/NIGMS NIH HHS/United States
- P30 EY008126/EY/NEI NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- CA68485/CA/NCI NIH HHS/United States
- DK20593/DK/NIDDK NIH HHS/United States
- U24 DK059637/DK/NIDDK NIH HHS/United States
- DK59637/DK/NIDDK NIH HHS/United States
- P30 DK020593/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

