Finding the fifth base: genome-wide sequencing of cytosine methylation
- PMID: 19273618
- PMCID: PMC3807530
- DOI: 10.1101/gr.083451.108
Finding the fifth base: genome-wide sequencing of cytosine methylation
Abstract
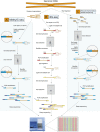
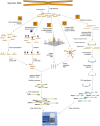
Complete sequences of myriad eukaryotic genomes, including several human genomes, are now available, and recent dramatic developments in DNA sequencing technology are opening the floodgates to vast volumes of sequence data. Yet, despite knowing for several decades that a significant proportion of cytosines in the genomes of plants and animals are present in the form of methylcytosine, until very recently the precise locations of these modified bases have never been accurately mapped throughout a eukaryotic genome. Advanced "next-generation" DNA sequencing technologies are now enabling the global mapping of this epigenetic modification at single-base resolution, providing new insights into the regulation and dynamics of DNA methylation in genomes.
Figures
References
-
- Absalan F., Ronaghi M. Molecular inversion probe assay. Methods Mol. Biol. 2007;396:315–330. - PubMed
-
- Albert T.J., Molla M.N., Muzny D.M., Nazareth L., Wheeler D., Song X., Richmond T.A., Middle C.M., Rodesch M.J., Packard C.J., et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–905. - PubMed
-
- Bashiardes S., Veile R., Helms C., Mardis E.R., Bowcock A.M., Lovett M. Direct genomic selection. Nat. Methods. 2005;2:63–69. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
