Concurrent titration and determination of antibiotic resistance in ureaplasma species with identification of novel point mutations in genes associated with resistance
- PMID: 19273669
- PMCID: PMC2681543
- DOI: 10.1128/AAC.01349-08
Concurrent titration and determination of antibiotic resistance in ureaplasma species with identification of novel point mutations in genes associated with resistance
Abstract
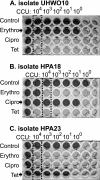
Antibiotic resistance determination of Ureaplasma spp. (Ureaplasma parvum and Ureaplasma urealyticum) usually requires predetermination of bacterial titer, followed by antibiotic interrogation using a set bacterial input. This 96-well method allows simultaneous quantification of bacteria in the presence and absence of antibiotics. A method for determining precise MICs and a method for screening against multiple antibiotics using breakpoint thresholds are detailed. Of the 61 Ureaplasma-positive clinical isolates screened, one (1.6%) was resistant to erythromycin (MIC, >64 mg/liter) and clarithromycin (MIC, 4 mg/liter), one to ciprofloxacin (1.6%), and one to tetracycline/doxycycline (1.6%). Five isolates were also consistently found to have an elevated MIC of 8 mg/liter for erythromycin, but this may not represent true antibiotic resistance, as no mutations were found in the 23S rRNA operons or ribosome-associated L4 and L22 proteins for these strains. However, two amino acids (R66Q67) were deleted from the L4 protein of the erythromycin-/clarithromycin-resistant strain. The tetM genetic element was detected in the tetracycline-resistant clinical isolate as well as in the positive control Vancouver strain serotype 9. The tetM gene was also found in a fully tetracycline-susceptible Ureaplasma clinical isolate, and no mutations were found in the coding region that would explain its failure to mediate tetracycline resistance. An amino acid substitution (D82N) was found in the ParC subunit of the ciprofloxacin-resistant isolate, adjacent to the S83L mutation reported by other investigators in many ciprofloxacin-resistant Ureaplasma isolates. It is now possible to detect antibiotic resistance in Ureaplasma within 48 h of positive culture without prior knowledge of bacterial load, identifying them for further molecular analysis.
Figures

Similar articles
-
Antibiotic susceptibilities and resistance genes of Ureaplasma parvum isolated in South Africa.J Antimicrob Chemother. 2012 Dec;67(12):2821-4. doi: 10.1093/jac/dks314. Epub 2012 Aug 9. J Antimicrob Chemother. 2012. PMID: 22879459
-
Antibiotic Susceptibility and Sequence Type Distribution of Ureaplasma Species Isolated from Genital Samples in Switzerland.Antimicrob Agents Chemother. 2015 Oct;59(10):6026-31. doi: 10.1128/AAC.00895-15. Epub 2015 Jul 20. Antimicrob Agents Chemother. 2015. PMID: 26195516 Free PMC article.
-
Antibacterial Resistance in Ureaplasma Species and Mycoplasma hominis Isolates from Urine Cultures in College-Aged Females.Antimicrob Agents Chemother. 2017 Sep 22;61(10):e01104-17. doi: 10.1128/AAC.01104-17. Print 2017 Oct. Antimicrob Agents Chemother. 2017. PMID: 28827422 Free PMC article.
-
Antibiotic resistance among Ureaplasma spp. isolates: cause for concern?J Antimicrob Chemother. 2017 Feb;72(2):330-337. doi: 10.1093/jac/dkw425. Epub 2016 Oct 20. J Antimicrob Chemother. 2017. PMID: 27798207 Review.
-
Clinical antibiotic resistance of Ureaplasma urealyticum.Pediatr Infect Dis. 1986 Nov-Dec;5(6 Suppl):S335-7. doi: 10.1097/00006454-198611010-00031. Pediatr Infect Dis. 1986. PMID: 3540906 Review.
Cited by
-
Regulation of tert-Butyl Hydroperoxide Resistance by Chromosomal OhrR in A. baumannii ATCC 19606.Microorganisms. 2021 Mar 18;9(3):629. doi: 10.3390/microorganisms9030629. Microorganisms. 2021. PMID: 33803549 Free PMC article.
-
Antimicrobial susceptibilities and mechanisms of resistance of commensal and invasive Mycoplasma salivarium isolates.Front Microbiol. 2022 Aug 1;13:914464. doi: 10.3389/fmicb.2022.914464. eCollection 2022. Front Microbiol. 2022. PMID: 35979479 Free PMC article.
-
Cervical epithelial damage promotes Ureaplasma parvum ascending infection, intrauterine inflammation and preterm birth induction in mice.Nat Commun. 2020 Jan 10;11(1):199. doi: 10.1038/s41467-019-14089-y. Nat Commun. 2020. PMID: 31924800 Free PMC article.
-
Incidence and antibiotic susceptibility of genital mycoplasmas in sexually active individuals in Hungary.Eur J Clin Microbiol Infect Dis. 2013 Nov;32(11):1423-6. doi: 10.1007/s10096-013-1892-y. Epub 2013 May 18. Eur J Clin Microbiol Infect Dis. 2013. PMID: 23686458
-
In vitro activity of five quinolones and analysis of the quinolone resistance-determining regions of gyrA, gyrB, parC, and parE in Ureaplasma parvum and Ureaplasma urealyticum clinical isolates from perinatal patients in Japan.Antimicrob Agents Chemother. 2015 Apr;59(4):2358-64. doi: 10.1128/AAC.04262-14. Epub 2015 Feb 2. Antimicrob Agents Chemother. 2015. PMID: 25645833 Free PMC article.
References
-
- Abele-Horn, M., J. Peters, O. Genzel-Boroviczeny, C. Wolff, A. Zimmermann, and W. Gottschling. 1997. Vaginal Ureaplasma urealyticum colonization: influence on pregnancy outcome and neonatal morbidity. Infection 25:286-291. - PubMed
-
- Bebear, C. M., H. Renaudin, A. Charron, D. Gruson, M. Lefrancois, and C. Bebear. 2000. In vitro activity of trovafloxacin compared to those of five antimicrobials against mycoplasmas including Mycoplasma hominis and Ureaplasma urealyticum fluoroquinolone-resistant isolates that have been genetically characterized. Antimicrob. Agents Chemother. 44:2557-2560. - PMC - PubMed
-
- Blanchard, A., D. M. Crabb, K. Dybvig, L. B. Duffy, and G. H. Cassell. 1992. Rapid detection of tetM in Mycoplasma hominis and Ureaplasma urealyticum by PCR: tetM confers resistance to tetracycline but not necessarily to doxycycline. FEMS Microbiol. Lett. 74:277-281. - PubMed
-
- Blanchard, A., J. Hentschel, L. Duffy, K. Baldus, and G. H. Cassell. 1993. Detection of Ureaplasma urealyticum by polymerase chain reaction in the urogenital tract of adults, in amniotic fluid, and in the respiratory tract of newborns. Clin. Infect. Dis. 17(Suppl. 1):S148-S153. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

