The interplay between transcription factors and microRNAs in genome-scale regulatory networks
- PMID: 19274664
- PMCID: PMC3118512
- DOI: 10.1002/bies.200800212
The interplay between transcription factors and microRNAs in genome-scale regulatory networks
Abstract
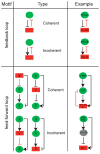
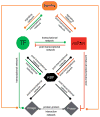
Metazoan genomes contain thousands of protein-coding and non-coding RNA genes, most of which are differentially expressed, i.e., at different locations, at different times during development, or in response to environmental signals. Differential gene expression is achieved through complex regulatory networks that are controlled in part by two types of trans-regulators: transcription factors (TFs) and microRNAs (miRNAs). TFs bind to cis-regulatory DNA elements that are often located in or near their target genes, while miRNAs hybridize to cis-regulatory RNA elements mostly located in the 3' untranslated region of their target mRNAs. Here, we describe how these trans-regulators interact with each other in the context of gene regulatory networks to coordinate gene expression at the genome-scale level, and discuss future challenges of integrating these networks with other types of functional networks.
Figures


References
-
- The C. elegans Sequencing Consortium, Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. - PubMed
-
- Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. - PubMed
-
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. - PubMed
-
- Levine M, Tjian R. Transcription regulation and animal diversity. Nature. 2003;424:147–151. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

