Structural analysis of the human cannabinoid receptor one carboxyl-terminus identifies two amphipathic helices
- PMID: 19274719
- PMCID: PMC2698224
- DOI: 10.1002/bip.21179
Structural analysis of the human cannabinoid receptor one carboxyl-terminus identifies two amphipathic helices
Abstract
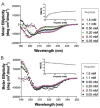
Recent research has implicated the C-terminus of G-protein coupled receptors in key events such as receptor activation and subsequent intracellular sorting, yet obtaining structural information of the entire C-tail has proven a formidable task. Here, a peptide corresponding to the full-length C-tail of the human CB1 receptor (residues 400-472) was expressed in E.coli and purified in a soluble form. Circular dichroism (CD) spectroscopy revealed that the peptide adopts an alpha-helical conformation in negatively charged and zwitterionic detergents (48-51% and 36-38%, respectively), whereas it exhibited the CD signature of unordered structure at low concentration in aqueous solution. Interestingly, 27% helicity was displayed at high peptide concentration suggesting that self-association induces helix formation in the absence of a membrane mimetic. NMR spectroscopy of the doubly labeled ((15)N- and (13)C-) C-terminus in dodecylphosphocholine (DPC) identified two amphipathic alpha-helical domains. The first domain, S401-F412, corresponds to the helix 8 common to G protein-coupled receptors while the second domain, A440-M461, is a newly identified structural motif in the distal region of the carboxyl-terminus of the receptor. Molecular modeling of the C-tail in DPC indicates that both helices lie parallel to the plane of the membrane with their hydrophobic and hydrophilic faces poised for critical interactions.
(c) 2009 Wiley Periodicals, Inc.
Figures


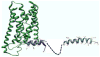


References
-
- Shire D, Carillon C, Kaghad M, Calandra B, Rinaldi-Carmona M, Le Fur G, Caput D, Ferrara P. J Biol Chem. 1995;270:3726–3731. - PubMed
-
- Ryberg E, Vu HK, Larsson N, Groblewski T, Hjorth S, Elebring T, Sjögren S, Greasley PJ. FEBS Lett. 2005;579:259–264. - PubMed
-
- Porter AC, Felder CC. Pharmacol Ther. 2001;90:45–60. - PubMed
-
- Ulfers AL, McMurry JL, Kendall DA, Mierke DF. Biochemistry. 2002;41:11344–11350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

