Campylobacter genotyping to determine the source of human infection
- PMID: 19275496
- PMCID: PMC3988352
- DOI: 10.1086/597402
Campylobacter genotyping to determine the source of human infection
Abstract
Background: Campylobacter species cause a high proportion of bacterial gastroenteritis cases and are a significant burden on health care systems and economies worldwide; however, the relative contributions of the various possible sources of infection in humans are unclear.
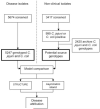
Methods: National-scale genotyping of Campylobacter species was used to quantify the relative importance of various possible sources of human infection. Multilocus sequence types were determined for 5674 isolates obtained from cases of human campylobacteriosis in Scotland from July 2005 through September 2006 and from 999 Campylobacter species isolates from 3417 contemporaneous samples from potential human infection sources. These data were supplemented with 2420 sequence types from other studies, representing isolates from a variety of sources. The clinical isolates were attributed to possible sources on the basis of their sequence types with use of 2 population genetic models, STRUCTURE and an asymmetric island model.
Results: The STRUCTURE and the asymmetric island models attributed most clinical isolates to chicken meat (58% and 78% of Campylobacter jejuni and 40% and 56% of Campylobacter coli isolates, respectively), identifying it as the principal source of Campylobacter infection in humans. Both models attributed the majority of the remaining isolates to ruminant sources, with relatively few isolates attributed to wild bird, environment, swine, and turkey sources.
Conclusions: National-scale genotyping was a practical and efficient methodology for the quantification of the contributions of different sources to human Campylobacter infection. Combined with the knowledge that retail chicken is routinely contaminated with Campylobacter, these results are consistent with the view that the largest reductions in human campylobacteriosis in industrialized countries will come from interventions that focus on the poultry industry.
Figures


References
-
- Oberhelman RA, Taylor DN. Campylobacter infections in developing countries. In: Nachamkin I, Blaser MJ, editors. Campylobacter. 2nd ed. ASM Press; Washington, DC: 2000. pp. 139–53.
-
- Olson CK, Ethelberg S, van Pelt W, Tauxe RV. Epidemiology of Campylobacter jejuni infections in industrialized nations. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. 3rd ed. ASM Press; Washington, DC: 2008. pp. 163–91.
-
- Allos B. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–6. - PubMed
-
- Kessel AS, Gillespie IA, O’Brien SJ, Adak GK, Humphrey TJ, Ward LR. General outbreaks of infectious intestinal disease linked with poultry, England and Wales, 1992-1999. Commun Dis Public Health. 2001;4:171–7. - PubMed
-
- Centers for Disease Control and Prevention (CDC) Division of Food-borne, Bacterial and Mycotic Diseases (DFBMD) listing. CDC; United States: 2008.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

