ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions
- PMID: 19275939
- PMCID: PMC4052679
- DOI: 10.1016/j.ymeth.2009.03.001
ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions
Abstract
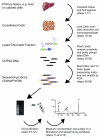
Chromatin immunoprecipitation (ChIP) allows specific protein-DNA interactions to be isolated. Combining ChIP with high-throughput sequencing reveals the DNA sequence involved in these interactions. Here, we describe how to perform ChIP-seq starting with whole tissues or cell lines, and ending with millions of short sequencing tags that can be aligned to the reference genome of the species under investigation. We also outline additional procedures to recover ChIP-chip libraries for ChIP-seq and discuss contemporary issues in data analysis.
Figures

References
-
- Elnitski L, Jin VX, Farnham PJ, Jones SJ. Locating mammalian transcription factor binding sites: a survey of computational and experimental techniques. Genome Res. 2006;16:1455–1464. - PubMed
-
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. - PubMed
-
- Cawley S, Bekiranov S, Ng HH, Kapranov P, Sekinger EA, et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

