The crystal structure of the signature domain of cartilage oligomeric matrix protein: implications for collagen, glycosaminoglycan and integrin binding
- PMID: 19276170
- PMCID: PMC2717772
- DOI: 10.1096/fj.08-128090
The crystal structure of the signature domain of cartilage oligomeric matrix protein: implications for collagen, glycosaminoglycan and integrin binding
Abstract
Cartilage oligomeric matrix protein (COMP), or thrombospondin-5 (TSP-5), is a secreted glycoprotein that is important for growth plate organization and function. Mutations in COMP cause two skeletal dysplasias, pseudoachondroplasia (PSACH) and multiple epiphyseal dysplasia (EDM1). In this study, we determined the structure of a recombinant protein that contains the last epidermal growth factor repeat, the type 3 repeats and the C-terminal domain (CTD) of COMP to 3.15-A resolution limit by X-ray crystallography. The CTD is a beta-sandwich that is composed of 15 antiparallel beta-strands, and the type 3 repeats are a contiguous series of calcium binding sites that associate with the CTD at multiple points. The crystal packing reveals an exposed potential metal-ion-dependent adhesion site (MIDAS) on one edge of the beta-sandwich that is common to all TSPs and may serve as a binding site for collagens and other ligands. Disease-causing mutations in COMP disrupt calcium binding, disulfide bond formation, intramolecular interactions, or sites for potential ligand binding. The structure presented here and its unique molecular packing in the crystal identify potential interactive sites for glycosaminoglycans, integrins, and collagens, which are key to cartilage structure and function.
Figures
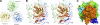


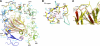

References
-
- Kipnes J, Carlberg A L, Loredo G A, Lawler J, Tuan R S, Hall D J. Effect of cartilage oligomeric matrix protein on mesenchymal chondrogenesis in vitro. Osteoarthritis Cartilage. 2003;11:442–454. - PubMed
-
- Xu K, Zhang Y, Ilalov K, Carlson C S, Feng J Q, Di Cesare P E, Liu C J. Cartilage oligomeric matrix protein associates with granulin-epithelin precursor (GEP) and potentiates GEP-stimulated chondrocyte proliferation. J Biol Chem. 2007;282:11347–11355. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

