Extracellular matrix-induced gene expression in human breast cancer cells
- PMID: 19276183
- PMCID: PMC2681181
- DOI: 10.1158/1541-7786.MCR-08-0227
Extracellular matrix-induced gene expression in human breast cancer cells
Abstract
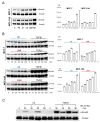
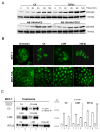
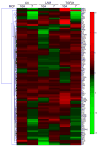
Extracellular matrix (ECM) molecules modify gene expression through attachment-dependent (focal adhesion-related) integrin receptor signaling. It was previously unknown whether the same molecules acting as soluble peptides could generate signal cascades without the associated mechanical anchoring, a condition that may be encountered during matrix remodeling and degradation and relevant to invasion and metastatic processes. In the current study, the role of ECM ligand-regulated gene expression through this attachment-independent process was examined. It was observed that fibronectin, laminin, and collagen type I and II induce Smad2 activation in MCF-10A and MCF-7 cells. This activation is not caused by transforming growth factor (TGF)-beta ligand contamination or autocrine TGF involvement and is 3- to 5-fold less robust than the TGF-beta1 ligand. The resulting nuclear translocation of Smad4 in response to ECM ligand indicates downstream transcriptional responses occurring. Coimmunoprecipitation experiments determined that collagen type II and laminin act through interaction with integrin alpha(2)beta(1) receptor complex. The ECM ligand-induced Smad activation (termed signaling crosstalk) resulted in cell type and ligand-specific transcriptional changes, which are distinct from the TGF-beta ligand-induced responses. These findings show that cell-matrix communication is more complex than previously thought. Soluble ECM peptides drive transcriptional regulation through corresponding adhesion and non-attachment-related processes. The resultant gene expressional patterns correlate with pathway activity and not by the extent of Smad activation. These results extend the complexity and the existing paradigms of ECM-cell communication to ECM ligand regulation without the necessity of mechanical coupling.
Figures




References
-
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. - PubMed
-
- Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2(6):442–54. - PubMed
-
- Atley LM, Mort JS, Lalumiere M, Eyre DR. Proteolysis of human bone collagen by cathepsin K: characterization of the cleavage sites generating by cross-linked N-telopeptide neoepitope. Bone. 2000;26(3):241–7. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

