Microfludic device for creating ionic strength gradients over DNA microarrays for efficient DNA melting studies and assay development
- PMID: 19277213
- PMCID: PMC2653225
- DOI: 10.1371/journal.pone.0004808
Microfludic device for creating ionic strength gradients over DNA microarrays for efficient DNA melting studies and assay development
Abstract
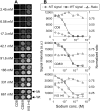
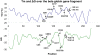
The development of DNA microarray assays is hampered by two important aspects: processing of the microarrays is done under a single stringency condition, and characteristics such as melting temperature are difficult to predict for immobilized probes. A technical solution to these limitations is to use a thermal gradient and information from melting curves, for instance to score genotypes. However, application of temperature gradients normally requires complicated equipment, and the size of the arrays that can be investigated is restricted due to heat dissipation. Here we present a simple microfluidic device that creates a gradient comprising zones of defined ionic strength over a glass slide, in which each zone corresponds to a subarray. Using this device, we demonstrated that ionic strength gradients function in a similar fashion as corresponding thermal gradients in assay development. More specifically, we noted that (i) the two stringency modulators generated melting curves that could be compared, (ii) both led to increased assay robustness, and (iii) both were associated with difficulties in genotyping the same mutation. These findings demonstrate that ionic strength stringency buffers can be used instead of thermal gradients. Given the flexibility of design of ionic gradients, these can be created over all types of arrays, and encompass an attractive alternative to temperature gradients, avoiding curtailment of the size or spacing of subarrays on slides associated with temperature gradients.
Conflict of interest statement
Figures
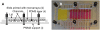



References
-
- Vainrub A, Pettitt BM. Surface electrostatic effects in oligonucleotide microarrays: control and optimization of binding thermodynamics. Biopolymers. 2003;68:265–270. - PubMed
-
- Marcy Y, Cousin PY, Rattier M, Cerovic G, Escalier G, et al. Innovative integrated system for real-time measurement of hybridization and melting on standard format microarrays. Biotechniques. 2008;44:913–920. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

