Control of high affinity interactions in the talin C terminus: how talin domains coordinate protein dynamics in cell adhesions
- PMID: 19278997
- PMCID: PMC2679484
- DOI: 10.1074/jbc.M900266200
Control of high affinity interactions in the talin C terminus: how talin domains coordinate protein dynamics in cell adhesions
Abstract
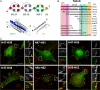
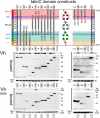
In cell-extracellular matrix junctions (focal adhesions), the cytoskeletal protein talin is central to the connection of integrins to the actin cytoskeleton. Talin is thought to mediate this connection via its two integrin, (at least) three actin, and several vinculin binding sites. The binding sites are cryptic in the head-to-rod autoinhibited cytoplasmic form of the protein and require (stepwise) conformational activation. This activation process, however, remains poorly understood, and there are contradictory models with respect to the determinants of adhesion site localization. Here, we report turnover rates and protein-protein interactions in a range of talin rod domain constructs varying in helix bundle structure. We conclude that several bundles of the C terminus cooperate to regulate targeting and concomitantly tailor high affinity interactions of the talin rod in cell adhesions. Intrinsic control of ligand binding activities is essential for the coordination of adhesion site function of talin.
Figures




Similar articles
-
The domain structure of talin: residues 1815-1973 form a five-helix bundle containing a cryptic vinculin-binding site.FEBS Lett. 2010 Jun 3;584(11):2237-41. doi: 10.1016/j.febslet.2010.04.028. Epub 2010 Apr 20. FEBS Lett. 2010. PMID: 20399778 Free PMC article.
-
Talin contains a C-terminal calpain2 cleavage site important in focal adhesion dynamics.PLoS One. 2012;7(4):e34461. doi: 10.1371/journal.pone.0034461. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496808 Free PMC article.
-
The Architecture of Talin1 Reveals an Autoinhibition Mechanism.Cell. 2019 Sep 19;179(1):120-131.e13. doi: 10.1016/j.cell.2019.08.034. Cell. 2019. PMID: 31539492 Free PMC article.
-
Integrin-mediated cell adhesion: the cytoskeletal connection.Biochem Soc Symp. 1999;65:79-99. Biochem Soc Symp. 1999. PMID: 10320934 Review.
-
Integrin connections to the cytoskeleton through talin and vinculin.Biochem Soc Trans. 2008 Apr;36(Pt 2):235-9. doi: 10.1042/BST0360235. Biochem Soc Trans. 2008. PMID: 18363566 Review.
Cited by
-
Selective Role of Vinculin in Contractile Mechanisms of Endothelial Permeability.Am J Respir Cell Mol Biol. 2016 Oct;55(4):476-486. doi: 10.1165/rcmb.2015-0328OC. Am J Respir Cell Mol Biol. 2016. PMID: 27115795 Free PMC article.
-
Cooperativity between integrin activation and mechanical stress leads to integrin clustering.Biophys J. 2011 Jun 8;100(11):2595-604. doi: 10.1016/j.bpj.2011.03.028. Biophys J. 2011. PMID: 21641304 Free PMC article.
-
Single and collective cell migration: the mechanics of adhesions.Mol Biol Cell. 2017 Jul 7;28(14):1833-1846. doi: 10.1091/mbc.E17-03-0134. Mol Biol Cell. 2017. PMID: 28684609 Free PMC article.
-
In vivo quantitative analysis of Talin turnover in response to force.Mol Biol Cell. 2015 Nov 5;26(22):4149-62. doi: 10.1091/mbc.E15-05-0304. Epub 2015 Oct 7. Mol Biol Cell. 2015. PMID: 26446844 Free PMC article.
-
Kank2 activates talin, reduces force transduction across integrins and induces central adhesion formation.Nat Cell Biol. 2016 Sep;18(9):941-53. doi: 10.1038/ncb3402. Epub 2016 Aug 22. Nat Cell Biol. 2016. PMID: 27548916 Free PMC article.
References
-
- Hynes, R. (2002) Cell 110 673-687 - PubMed
-
- Liu, S., Calderwood, D. A., and Ginsberg, M. H. (2000) J. Cell Sci. 113 3563-3571 - PubMed
-
- Jiang, G., Giannone, G., Critchley, D. R., Fukumoto, E., and Sheetz, M. P. (2003) Nature 424 334-337 - PubMed
-
- Tadokoro, S., Shattil, S. J., Eto, K., Tai, V., Liddington, R. C., de Pereda, J. M., Ginsberg, M. H., and Calderwood, D. A. (2003) Science 302 103-106 - PubMed
-
- Nayal, A., Webb, D. J., and Horwitz, A. F. (2004) Curr. Opin. Cell Biol. 16 1-5 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

