Cell wall beta-(1,6)-glucan of Saccharomyces cerevisiae: structural characterization and in situ synthesis
- PMID: 19279004
- PMCID: PMC2679440
- DOI: 10.1074/jbc.M807667200
Cell wall beta-(1,6)-glucan of Saccharomyces cerevisiae: structural characterization and in situ synthesis
Abstract
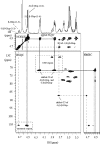
Despite its essential role in the yeast cell wall, the exact composition of the beta-(1,6)-glucan component is not well characterized. While solubilizing the cell wall alkali-insoluble fraction from a wild type strain of Saccharomyces cerevisiae using a recombinant beta-(1,3)-glucanase followed by chromatographic characterization of the digest on an anion exchange column, we observed a soluble polymer that eluted at the end of the solvent gradient run. Further characterization indicated this soluble polymer to have a molecular mass of approximately 38 kDa and could be hydrolyzed only by beta-(1,6)-glucanase. Gas chromatography mass spectrometry and NMR ((1)H and (13)C) analyses confirmed it to be a beta-(1,6)-glucan polymer with, on average, branching at every fifth residue with one or two beta-(1,3)-linked glucose units in the side chain. This polymer peak was significantly reduced in the corresponding digests from mutants of the kre genes (kre9 and kre5) that are known to play a crucial role in the beta-(1,6)-glucan biosynthesis. In the current study, we have developed a biochemical assay wherein incubation of UDP-[(14)C]glucose with permeabilized S. cerevisiae yeasts resulted in the synthesis of a polymer chemically identical to the branched beta-(1,6)-glucan isolated from the cell wall. Using this assay, parameters essential for beta-(1,6)-glucan synthetic activity were defined.
Figures

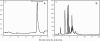



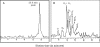

References
-
- Kapteyn, J. C., Van Den Ende, H., and Klis, F. M. (1999) Biochim. Biophys. Acta 1426 373–383 - PubMed
-
- Kollar, R., Reinhold, B. B., Petrakova, E., Yeh, H. J., Ashwell, G., Drgonova, J., Kapteyn, J. C., Klis, F. M., and Cabib, E. (1997) J. Biol. Chem. 272 17762–17775 - PubMed
-
- Humbel, B. M., Konomi, M., Takagi, T., Kamasawa, N., Ishijima, S. A., and Osumi, M. (2001) Yeast 18 433–444 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

