GATA transcription factors regulate the expression of the human eosinophil-derived neurotoxin (RNase 2) gene
- PMID: 19279013
- PMCID: PMC2676042
- DOI: 10.1074/jbc.M807307200
GATA transcription factors regulate the expression of the human eosinophil-derived neurotoxin (RNase 2) gene
Abstract
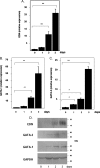
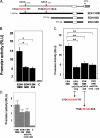
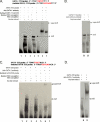
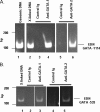
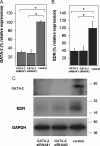
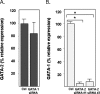
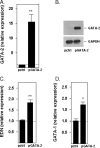
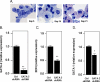
The transcription factors GATA-1 and GATA-2 have been implicated in promoting differentiation of eosinophilic leukocytes. In this study, we examined the roles of GATA-1 and GATA-2 in activating transcription of the secretory ribonuclease, the eosinophil-derived neurotoxin (EDN/RNase 2). Augmented expression of both GATA-1 and GATA-2 was detected in eosinophil promyelocyte HL-60 clone 15 cells in response to biochemical differentiation with butyric acid. Deletion or mutation of one or both of the two consensus GATA-binding sites in the extended 1000-bp 5' promoter of the EDN gene resulted in profound reduction in reporter gene activity. Antibody-augmented electrophoretic mobility shift and chromatin immunoprecipitation analyses indicate that GATA-1 and GATA-2 proteins bind to both functional GATA consensus sequences in the EDN promoter. Interestingly, RNA silencing of GATA-1 alone had no impact on EDN expression; silencing of GATA-2 resulted in diminished expression of EDN, and also diminished expression of GATA-1 in both butyric acid-induced HL-60 clone 15 cells and in differentiating human eosinophils derived from CD34(+) hematopoietic progenitors. Likewise, overexpression of GATA-2 in uninduced HL-60 clone 15 cells resulted in augmented transcription of both EDN and GATA-1. Taken together, our data suggest that GATA-2 functions directly via interactions with the EDN promoter and also indirectly, via its ability to regulate the expression of GATA-1 in differentiating eosinophils and eosinophil cell lines.
Figures

Similar articles
-
Transcriptional regulation of human eosinophil RNases by an evolutionary- conserved sequence motif in primate genome.BMC Mol Biol. 2007 Oct 11;8:89. doi: 10.1186/1471-2199-8-89. BMC Mol Biol. 2007. PMID: 17927842 Free PMC article.
-
Enhanced expression of the eosinophil-derived neurotoxin ribonuclease (RNS2) gene requires interaction between the promoter and intron.J Biol Chem. 1996 May 24;271(21):12387-93. doi: 10.1074/jbc.271.21.12387. J Biol Chem. 1996. PMID: 8647842
-
Transcriptional regulation of human eosinophil RNase2 by the liver-enriched hepatocyte nuclear factor 4.J Cell Biochem. 2009 Feb 1;106(2):317-26. doi: 10.1002/jcb.22008. J Cell Biochem. 2009. PMID: 19115260
-
Eosinophil-Derived Neurotoxin (EDN/RNase 2) and the Mouse Eosinophil-Associated RNases (mEars): Expanding Roles in Promoting Host Defense.Int J Mol Sci. 2015 Jul 8;16(7):15442-55. doi: 10.3390/ijms160715442. Int J Mol Sci. 2015. PMID: 26184157 Free PMC article. Review.
-
Eosinophil-derived neurotoxin / RNase 2: connecting the past, the present and the future.Curr Pharm Biotechnol. 2008 Jun;9(3):135-40. doi: 10.2174/138920108784567236. Curr Pharm Biotechnol. 2008. PMID: 18673278 Free PMC article. Review.
Cited by
-
The Immunomodulatory and Antimicrobial Properties of the Vertebrate Ribonuclease A Superfamily.Vaccines (Basel). 2018 Nov 20;6(4):76. doi: 10.3390/vaccines6040076. Vaccines (Basel). 2018. PMID: 30463297 Free PMC article. Review.
-
A CREB-mediated increase in miRNA let-7f during prolonged β-agonist exposure: a novel mechanism of β2-adrenergic receptor down-regulation in airway smooth muscle.FASEB J. 2018 Jul;32(7):3680-3688. doi: 10.1096/fj.201701278R. Epub 2018 Feb 13. FASEB J. 2018. PMID: 29455573 Free PMC article.
-
Bexarotene inhibits the viability of non-small cell lung cancer cells via slc10a2/PPARγ/PTEN/mTOR signaling pathway.BMC Cancer. 2018 Apr 11;18(1):407. doi: 10.1186/s12885-018-4224-x. BMC Cancer. 2018. PMID: 29642873 Free PMC article.
-
The gene structure and promoter region of the vaccine target aminopeptidase H11 from the blood-sucking nematode parasite of ruminants, Haemonchus contortus.Funct Integr Genomics. 2010 Nov;10(4):589-601. doi: 10.1007/s10142-010-0172-5. Epub 2010 May 1. Funct Integr Genomics. 2010. PMID: 20437190
-
RhoH is a negative regulator of eosinophilopoiesis.Cell Death Differ. 2016 Dec;23(12):1961-1972. doi: 10.1038/cdd.2016.73. Epub 2016 Oct 14. Cell Death Differ. 2016. PMID: 27740624 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

