Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase
- PMID: 19279203
- PMCID: PMC2654394
- DOI: 10.1073/pnas.0901403106
Translesion DNA polymerases remodel the replisome and alter the speed of the replicative helicase
Abstract
All cells contain specialized translesion DNA polymerases that replicate past sites of DNA damage. We find that Escherichia coli translesion DNA polymerase II (Pol II) and polymerase IV (Pol IV) function with DnaB helicase and regulate its rate of unwinding, slowing it to as little as 1 bp/s. Furthermore, Pol II and Pol IV freely exchange with the polymerase III (Pol III) replicase on the beta-clamp and function with DnaB helicase to form alternative replisomes, even before Pol III stalls at a lesion. DNA damage-induced levels of Pol II and Pol IV dominate the clamp, slowing the helicase and stably maintaining the architecture of the replication machinery while keeping the fork moving. We propose that these dynamic actions provide additional time for normal excision repair of lesions before the replication fork reaches them and also enable the appropriate translesion polymerase to sample each lesion as it is encountered.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


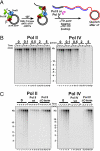
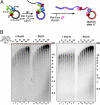
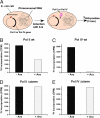
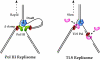
Comment in
-
Shifting replication between IInd, IIIrd, and IVth gears.Proc Natl Acad Sci U S A. 2009 Apr 14;106(15):6027-8. doi: 10.1073/pnas.0902226106. Epub 2009 Apr 8. Proc Natl Acad Sci U S A. 2009. PMID: 19357302 Free PMC article. No abstract available.
References
-
- Kornberg A, Baker TA. DNA Replication. 2nd ed. New York: Freeman Press; 1992. DNA replication; p. 931.
-
- Johnson A, O'Donnell M. Cellular DNA replicases: Components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. - PubMed
-
- Kim S, Dallmann HG, McHenry CS, Marians KJ. Coupling of a replicative polymerase and helicase: A tau–DnaB interaction mediates rapid replication fork movement. Cell. 1996;84:643–650. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

