Binding to gating transduction in nicotinic receptors: Cys-loop energetically couples to pre-M1 and M2-M3 regions
- PMID: 19279256
- PMCID: PMC2728446
- DOI: 10.1523/JNEUROSCI.6185-08.2009
Binding to gating transduction in nicotinic receptors: Cys-loop energetically couples to pre-M1 and M2-M3 regions
Abstract
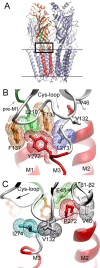
The nicotinic acetylcholine receptor (AChR) transduces binding of nerve-released ACh into opening of an intrinsic ion channel, yet the intraprotein interactions behind transduction remain to be fully elucidated. Attention has focused on the region of the AChR in which the beta1-beta2 and Cys-loops from the extracellular domain project into a cavity framed by residues preceding the first transmembrane domain (pre-M1) and the linker spanning transmembrane domains M2 and M3. Previous studies identified a principal transduction pathway in which the pre-M1 domain is coupled to the M2-M3 linker through the beta1-beta2 loop. Here we identify a parallel pathway in which the pre-M1 domain is coupled to the M2-M3 linker through the Cys-loop. Mutagenesis, single-channel kinetic analyses and thermodynamic mutant cycle analyses reveal energetic coupling among alphaLeu 210 from the pre-M1 domain, alphaPhe 135 and alphaPhe 137 from the Cys-loop, and alphaLeu 273 from the M2-M3 linker. Residues at equivalent positions of non-alpha-subunits show negligible coupling, indicating these interresidue couplings are specific to residues in the alpha-subunit. Thus, the extracellular beta1-beta2 and Cys-loops bridge the pre-M1 domain and M2-M3 linker to transduce agonist binding into channel gating.
Figures





References
-
- Albeck S, Unger R, Schreiber G. Evaluation of direct and cooperative contributions towards the strength of buried hydrogen bonds and salt bridges. J Mol Biol. 2000;298:503–520. - PubMed
-
- Armstrong N, Sun Y, Chen GQ, Gouaux E. Structure of a glutamate-receptor ligand-binding core in complex with kainate. Nature. 1998;395:913–917. - PubMed
-
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. - PubMed
-
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. - PubMed
-
- Carter PJ, Winter G, Wilkinson AJ, Fersht AR. The use of double mutants to detect structural changes in the active site of the tyrosyl-tRNA synthetase (Bacillus stearothermophilus) Cell. 1984;38:835–840. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
