In vivo and in vitro differentiation of uniparental embryonic stem cells into hematopoietic and neural cell types
- PMID: 19279713
- PMCID: PMC2634177
- DOI: 10.4161/org.6123
In vivo and in vitro differentiation of uniparental embryonic stem cells into hematopoietic and neural cell types
Abstract
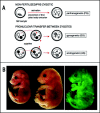
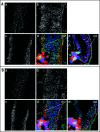
The biological role of genomic imprinting in adult tissue is central to the consideration of transplanting uniparental embryonic stem (ES) cell-derived tissues. We have recently shown that both maternal (parthenogenetic/gynogenetic) and paternal (androgenetic) uniparental ES cells can differentiate, both in vivo in chimeras and in vitro, into adult-repopulating hematopoietic stem and progenitor cells. This suggests that, at least in some tissues, the presence of two maternal or two paternal genomes does not interfere with stem cell function and tissue homeostasis in the adult. Here, we consider implications of the contribution of uniparental cells to hematopoiesis and to development of other organ systems, notably neural tissue for which consequences of genomic imprinting are associated with a known bias in development and behavioral disorders. Our findings so far indicate that there is little or no limit to the differentiation potential of uniparental ES cells outside the normal developmental paradigm. As a potentially donor MHC-matching source of tissue, uniparental transplants may provide not only a clinical resource but also a unique tool to investigate aspects of genomic imprinting in adults.
Keywords: androgenetic; chimera; gynogenetic; hematopoietic; neural; parthenogenetic; transplantation; uniparental.
Figures




References
-
- Surani MA, Barton SC, Norris ML. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. - PubMed
-
- McGrath J, Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. - PubMed
-
- Cattanach BM, Kirk M. Differential activity of maternally and paternally derived chromosome regions in mice. Nature. 1985;315:496–498. - PubMed
-
- Barlow DP, Stoger R, Herrmann BG, Saito K, Schweifer N. The mouse insulin-like growth factor type-2 receptor is imprinted and closely linked to the Tme locus. Nature. 1991;349:84–87. - PubMed
-
- DeChiara TM, Robertson EJ, Efstratiadis A. Parental imprinting of the mouse insulin-like growth factor II gene. Cell. 1991;64:849–859. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
