Gene content, organization and molecular evolution of plant organellar genomes and sex chromosomes: insights from the case of the liverwort Marchantia polymorpha
- PMID: 19282647
- PMCID: PMC3524301
- DOI: 10.2183/pjab.85.108
Gene content, organization and molecular evolution of plant organellar genomes and sex chromosomes: insights from the case of the liverwort Marchantia polymorpha
Abstract
The complete nucleotide sequence of chloroplast DNA (121,025 base pairs, bp) from a liverwort, Marchantia polymorpha, has made clear the entire gene organization of the chloroplast genome. Quite a few genes encoding components of photosynthesis and protein synthesis machinery have been identified by comparative computer analysis. We also determined the complete nucleotide sequence of the liverwort mitochondrial DNA and deduced 96 possible genes in the sequence of 186,608 bp. The complete chloroplast genome encodes twenty introns (19 group II and 1 group I) in 18 different genes. One of the chloroplast group II introns separates a ribosomal protein gene in a trans-position. The mitochondrial genome contains thirty-two introns (25 group II and 7 group I) in the coding regions of 17 genes. From the evolutionary point of view, we describe the origin of organellar introns and give evidence for vertical and horizontal intron transfers and their intragenomic propagation. Furthermore, we describe the gene organization of the Y chromosome in the dioecious liverwort M. polymorpha, the first detailed view of a Y chromosome in a haploid organism. On the 10 megabase (Mb) Y chromosome, 64 genes are identified, 14 of which are detected only in the male genome. These 14 genes are expressed in reproductive organs but not in vegetative thalli, suggesting their participation in male reproductive functions. These findings indicate that the Y and X chromosomes share the same ancestral autosome and support the prediction that in a haploid organism essential genes on sex chromosomes are more likely to persist than in a diploid organism.
Figures

 Photosynthesis and electron transport (rbc, psa, psb, pet, ndh, atp, frx);
Photosynthesis and electron transport (rbc, psa, psb, pet, ndh, atp, frx);
 Transcription (rpo);
Transcription (rpo);
 Translation (rpl, rps, rrn, trn);
Translation (rpl, rps, rrn, trn);
 Miscelleaneous (mbp, chl, clp);
Miscelleaneous (mbp, chl, clp);
 Unidentified ORF.–
Unidentified ORF.–
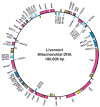
 Respiration (cox, cob, nad, atp);
Respiration (cox, cob, nad, atp);
 Translation (rpl, rps, rrn, trn);
Translation (rpl, rps, rrn, trn);
 Miscelleaneous (sdh, ccb, mtt);
Miscelleaneous (sdh, ccb, mtt);
 Unidentified ORF.,
Unidentified ORF.,




Similar articles
-
Chloroplast and mitochondrial genomes from a liverwort, Marchantia polymorpha--gene organization and molecular evolution.Biosci Biotechnol Biochem. 1996 Jan;60(1):16-24. doi: 10.1271/bbb.60.16. Biosci Biotechnol Biochem. 1996. PMID: 8824820 Review.
-
Gene organization of the liverwort Y chromosome reveals distinct sex chromosome evolution in a haploid system.Proc Natl Acad Sci U S A. 2007 Apr 10;104(15):6472-7. doi: 10.1073/pnas.0609054104. Epub 2007 Mar 29. Proc Natl Acad Sci U S A. 2007. PMID: 17395720 Free PMC article.
-
Identification of the sex-determining factor in the liverwort Marchantia polymorpha reveals unique evolution of sex chromosomes in a haploid system.Curr Biol. 2021 Dec 20;31(24):5522-5532.e7. doi: 10.1016/j.cub.2021.10.023. Epub 2021 Nov 3. Curr Biol. 2021. PMID: 34735792 Free PMC article.
-
Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification.J Mol Biol. 1988 Sep 20;203(2):281-98. doi: 10.1016/0022-2836(88)90001-0. J Mol Biol. 1988. PMID: 2462054
-
MicroRNAs in Marchantia polymorpha.New Phytol. 2018 Oct;220(2):409-416. doi: 10.1111/nph.15294. Epub 2018 Jun 30. New Phytol. 2018. PMID: 29959894 Review.
Cited by
-
A Cyan Fluorescent Reporter Expressed from the Chloroplast Genome of Marchantia polymorpha.Plant Cell Physiol. 2016 Feb;57(2):291-9. doi: 10.1093/pcp/pcv160. Epub 2015 Dec 3. Plant Cell Physiol. 2016. PMID: 26634291 Free PMC article.
-
chlB requirement for chlorophyll biosynthesis under short photoperiod in Marchantia polymorpha L.Genome Biol Evol. 2014 Mar;6(3):620-8. doi: 10.1093/gbe/evu045. Genome Biol Evol. 2014. PMID: 24586029 Free PMC article.
-
Nuclear DYW-type PPR gene families diversify with increasing RNA editing frequencies in liverwort and moss mitochondria.J Mol Evol. 2012 Feb;74(1-2):37-51. doi: 10.1007/s00239-012-9486-3. J Mol Evol. 2012. PMID: 22302222
-
Synthetic Botany.Cold Spring Harb Perspect Biol. 2017 Jul 5;9(7):a023887. doi: 10.1101/cshperspect.a023887. Cold Spring Harb Perspect Biol. 2017. PMID: 28246181 Free PMC article. Review.
-
Organellar genome assembly methods and comparative analysis of horticultural plants.Hortic Res. 2018 Jan 10;5:3. doi: 10.1038/s41438-017-0002-1. eCollection 2018. Hortic Res. 2018. PMID: 29423233 Free PMC article.
References
-
- Whitfeld P.R., Bottomley W. (1983) Organization and structure of chloroplast genes. Annu. Rev. Plant Physiol. 34, 279–310
-
- Palmer J.D. (1985) Comparative organization of chloroplast genomes. Annu. Rev. Genet. 19, 325–354 - PubMed
-
- Ohyama K., Fukuzawa H., Kohchi T., Shirai H., Sano T., Sano S., et al. (1986) Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322, 572–574
-
- Ohyama K., Fukuzawa H., Kohchi T., Sano T., Sano S., Shirai H., et al. (1988) Structure and organization of Marchantia polymorpha chloroplast genome. I. Cloning and gene identification. J. Mol. Biol. 203, 281–298 - PubMed
-
- Ohyama K., Kohchi T., Fukuzawa H., Sano T., Umesono K., Ozeki H. (1988) Gene organization and newly identified groups of genes of the chloroplast genome from a liverwort, Marchantia polymorpha. Photosynth. Res. 16, 7–22 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

