Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events
- PMID: 19282980
- PMCID: PMC2650098
- DOI: 10.1371/journal.ppat.1000340
Definition of herpes simplex virus type 1 helper activities for adeno-associated virus early replication events
Abstract
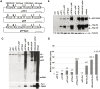
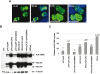
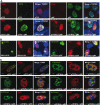
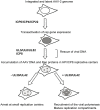
The human parvovirus Adeno-Associated Virus (AAV) type 2 can only replicate in cells co-infected with a helper virus, such as Adenovirus or Herpes Simplex Virus type 1 (HSV-1); whereas, in the absence of a helper virus, it establishes a latent infection. Previous studies demonstrated that the ternary HSV-1 helicase/primase (HP) complex (UL5/8/52) and the single-stranded DNA-Binding Protein (ICP8) were sufficient to induce AAV-2 replication in transfected cells. We independently showed that, in the context of a latent AAV-2 infection, the HSV-1 ICP0 protein was able to activate rep gene expression. The present study was conducted to integrate these observations and to further explore the requirement of other HSV-1 proteins during early AAV replication steps, i.e. rep gene expression and AAV DNA replication. Using a cellular model that mimics AAV latency and composite constructs coding for various sets of HSV-1 genes, we first confirmed the role of ICP0 for rep gene expression and demonstrated a synergistic effect of ICP4 and, to a lesser extent, ICP22. Conversely, ICP27 displayed an inhibitory effect. Second, our analyses showed that the effect of ICP0, ICP4, and ICP22 on rep gene expression was essential for the onset of AAV DNA replication in conjunction with the HP complex and ICP8. Third, and most importantly, we demonstrated that the HSV-1 DNA polymerase complex (UL30/UL42) was critical to enhance AAV DNA replication to a significant level in transfected cells and that its catalytic activity was involved in this process. Altogether, this work represents the first comprehensive study recapitulating the series of early events taking place during HSV-1-induced AAV replication.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Berns KI, Giraud C. Biology of adeno-associated virus. Curr Top Microbiol Immunol. 1996;218:1–23. - PubMed
-
- Muzyczka N, Berns KI. Parvoviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia, PA: Lippincott Williams & Wilkins; 2001. pp. 2327–2359.
-
- Geoffroy MC, Salvetti A. Helper functions required for wild type and recombinant adeno-associated virus growth. Curr Gene Ther. 2005;5:265–271. - PubMed
-
- Mishra L, Rose JA. Adeno-associated virus DNA replication is induced by genes that are essential for HSV-1 DNA synthesis. Virology. 1990;179:632–639. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

