Aphids acquired symbiotic genes via lateral gene transfer
- PMID: 19284544
- PMCID: PMC2662799
- DOI: 10.1186/1741-7007-7-12
Aphids acquired symbiotic genes via lateral gene transfer
Abstract
Background: Aphids possess bacteriocytes, which are cells specifically differentiated to harbour the obligate mutualist Buchnera aphidicola (gamma-Proteobacteria). Buchnera has lost many of the genes that appear to be essential for bacterial life. From the bacteriocyte of the pea aphid Acyrthosiphon pisum, we previously identified two clusters of expressed sequence tags that display similarity only to bacterial genes. Southern blot analysis demonstrated that they are encoded in the aphid genome. In this study, in order to assess the possibility of lateral gene transfer, we determined the full-length sequences of these transcripts, and performed detailed structural and phylogenetic analyses. We further examined their expression levels in the bacteriocyte using real-time quantitative RT-PCR.
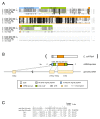
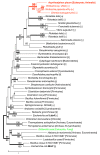
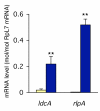
Results: Sequence similarity searches demonstrated that these fully sequenced transcripts are significantly similar to the bacterial genes ldcA (product, LD-carboxypeptidase) and rlpA (product, rare lipoprotein A), respectively. Buchnera lacks these genes, whereas many other bacteria, including Escherichia coli, a close relative of Buchnera, possess both ldcA and rlpA. Molecular phylogenetic analysis clearly demonstrated that the aphid ldcA was derived from a rickettsial bacterium closely related to the extant Wolbachia spp. (alpha-Proteobacteria, Rickettsiales), which are intracellular symbionts of various lineages of arthropods. The evolutionary origin of rlpA was not fully resolved, but it was clearly demonstrated that its double-psi beta-barrel domain is of bacterial origin. Real-time quantitative RT-PCR demonstrated that ldcA and rlpA are expressed 11.6 and 154-fold higher in the bacteriocyte than in the whole body, respectively. LdcA is an enzyme required for recycling murein (peptidoglycan), which is a component of the bacterial cell wall. As Buchnera possesses a cell wall composed of murein but lacks ldcA, a high level of expression of the aphid ldcA in the bacteriocyte may be essential to maintain Buchnera. Although the function of RlpA is not well known, conspicuous up-regulation of the aphid rlpA in the bacteriocyte implies that this gene is also essential for Buchnera.
Conclusion: In this study, we obtained several lines of evidence indicating that aphids acquired genes from bacteria via lateral gene transfer and that these genes are used to maintain the obligately mutualistic bacterium, Buchnera.
Figures


Comment in
-
Lateral gene transfer between prokaryotes and multicellular eukaryotes: ongoing and significant?BMC Biol. 2009 May 5;7:20. doi: 10.1186/1741-7007-7-20. BMC Biol. 2009. PMID: 19416510 Free PMC article.
References
-
- Buchner P. Endosymbiosis of animals with plant microorganisms. New York: Interscience; 1965.
-
- Munson MA, Baumann P, Kinsey MG. Buchnera gen. nov. and Buchnera aphidicola sp. nov., a taxon consisting of the mycetocyte-associated, primary endosymbionts of aphids. Int J Syst Bacteriol. 1991;41:566–568.
-
- Moran NA, Munson MA, Baumann P, Ishikawa H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. P Roy Soc Lond B Bio. 1993;253:167–171. doi: 10.1098/rspb.1993.0098. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

