The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods
- PMID: 19284646
- PMCID: PMC2680895
- DOI: 10.1186/1471-2164-10-107
The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods
Abstract
Background: The apparent scarcity of available sequence data has greatly impeded evolutionary studies in Acari (mites and ticks). This subclass encompasses over 48,000 species and forms the largest group within the Arachnida. Although mitochondrial genomes are widely utilised for phylogenetic and population genetic studies, only 20 mitochondrial genomes of Acari have been determined, of which only one belongs to the diverse order of the Sarcoptiformes. In this study, we describe the mitochondrial genome of the European house dust mite Dermatophagoides pteronyssinus, the most important member of this largely neglected group.
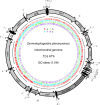
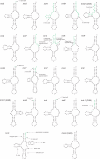
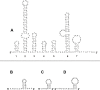
Results: The mitochondrial genome of D. pteronyssinus is a circular DNA molecule of 14,203 bp. It contains the complete set of 37 genes (13 protein coding genes, 2 rRNA genes and 22 tRNA genes), usually present in metazoan mitochondrial genomes. The mitochondrial gene order differs considerably from that of other Acari mitochondrial genomes. Compared to the mitochondrial genome of Limulus polyphemus, considered as the ancestral arthropod pattern, only 11 of the 38 gene boundaries are conserved. The majority strand has a 72.6% AT-content but a GC-skew of 0.194. This skew is the reverse of that normally observed for typical animal mitochondrial genomes. A microsatellite was detected in a large non-coding region (286 bp), which probably functions as the control region. Almost all tRNA genes lack a T-arm, provoking the formation of canonical cloverleaf tRNA-structures, and both rRNA genes are considerably reduced in size. Finally, the genomic sequence was used to perform a phylogenetic study. Both maximum likelihood and Bayesian inference analysis clustered D. pteronyssinus with Steganacarus magnus, forming a sistergroup of the Trombidiformes.
Conclusion: Although the mitochondrial genome of D. pteronyssinus shares different features with previously characterised Acari mitochondrial genomes, it is unique in many ways. Gene order is extremely rearranged and represents a new pattern within the Acari. Both tRNAs and rRNAs are truncated, corroborating the theory of the functional co-evolution of these molecules. Furthermore, the strong and reversed GC- and AT-skews suggest the inversion of the control region as an evolutionary event. Finally, phylogenetic analysis using concatenated mt gene sequences succeeded in recovering Acari relationships concordant with traditional views of phylogeny of Acari.
Figures




References
-
- Harvey MS. The neglected cousins: What do we know about the smaller Arachnid orders? J Arachnol. 2002;30:357–372.
-
- Acari. The Mites http://tolweb.org/Acari/2554/1996.12.13
-
- Dunlop JA, Alberti G. The affinities of mites and ticks: a review. J Zool Syst Evol Res. 2008;46:1–18.
-
- O'Connor BM. Phylogenetic relationships among higher taxa in the Acariformes, with particular reference to the Astigmata. In: Griffiths DA, Bowman CE, editor. Acarology VI. Vol. 1. Chichester: Ellis-Horwood Ltd; 1984. pp. 19–27.
-
- Spieksma FTM. Domestic mites from an acarologic perspectives. Allergy. 1997;52:360–368. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

