Characterization of the epithelial sodium channel alpha subunit coding and non-coding transcripts and their corresponding mRNA expression levels in Dahl R versus S rat kidney cortex on normal and high salt diet
- PMID: 19284664
- PMCID: PMC2669474
- DOI: 10.1186/1755-7682-2-5
Characterization of the epithelial sodium channel alpha subunit coding and non-coding transcripts and their corresponding mRNA expression levels in Dahl R versus S rat kidney cortex on normal and high salt diet
Abstract
Aims/hypothesis: The alpha subunit of the amiloride-sensitive epithelial sodium channel (alpha ENaC) is critical for the expression of functional channels. In humans and rats, non functional alternatively spliced forms of alpha ENaC have been proposed to act as negative regulatory components for ENaC. The purpose of this study was to examine the presence and consequently investigate the mRNA expression levels of alternatively spliced forms of alpha ENaC in kidney cortex of Dahl salt-resistant rats (R) versus Dahl salt-sensitive rats (S) on high salt and normal diets.
Methods: Using quantitative RT-PCR strategy, we examined the mRNA expression levels of previously reported alpha ENaC-a and -b alternatively spliced forms in kidney cortex of Dahl S and R rats on normal and four-week high salt diet and compared their corresponding abundance to wildtype alpha ENaC mRNA levels. We identified 2 novel non-coding C-terminus spliced forms and examined their mRNA expression in Dahl R versus S rat kidney cortex. We also tested the presence of five previously reported lung-specific alpha ENaC spliced forms in Dahl rat kidney cortex (CK479583, CK475461, CK364785, CK475819, and CB690980).
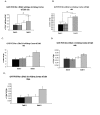
Results: Previously reported alpha ENaC-a and -b alternatively spliced forms are present in Dahl rat kidney cortex and are significantly higher in Dahl R versus S rats (P < 0.05). Four-week high salt diet significantly increases alpha ENaC-b (P < 0.05), but not alpha ENaC-a transcript abundance in Dahl R, but not S rats. Two non-coding alpha ENaC spliced forms -c and -d are newly identified in the present study, whose levels are comparable in Dahl R and S rats. Compared to alpha ENaC-wt, alpha ENaC-a, -c and -d are low abundance transcripts (4 +/- 2, 110 +/- 20, and 10 +/- 2 fold less respectively), in contrast to alpha ENaC-b abundance that exceeds alpha ENaC-wt by 32 +/- 3 fold. We could not identify any of the five previously reported lung-specific alpha ENaC spliced forms (CK479583, CK475461, CK364785, CK475819, and CB690980) in Dahl rat kidney cortex.
Conclusion/interpretation: alpha ENaC alternative splicing might regulate alpha ENaC by the formation of coding RNA species (alpha ENaC-a and -b) and non-coding RNA species (alpha ENaC-c and -d). alpha ENaC-a and -b mRNA levels are significantly higher in Dahl R versus S rats. Additionally, alpha ENaC-b is a salt-sensitive transcript whose levels are significantly higher 4-weeks post high salt diet compared to normal salt diet in Dahl R rats. Among the four alpha ENaC transcripts (-a, -b, -c and -d), alpha ENaC-b is a predominant transcript that exceeds alpha ENaC-wt abundance by ~32 fold. alpha ENaC-a and -b spliced forms, particularly, alpha ENaC-b, might potentially act as dominant negative proteins for ENaC activity, thereby rescuing Dahl R rats from developing salt-sensitive hypertension on high salt diet. On the other hand, non-coding alpha ENaC-c and -d might assist alternative splicing, facilitate RNA processing, or regulate alpha ENaC as well as each other.
Figures


References
-
- Garty H, Benos DJ. Characteristics and regulatory mechanisms of the amiloride-blockable Na+ channel. Physiol Rev. 1988;68:309–373. - PubMed
-
- Benos DJ, Saccomani G, Sariban-Sohraby S. The epithelial sodium channel. Subunit number and location of the amiloride binding site. J Biol Chem. 1987;262:10613–10618. - PubMed
LinkOut - more resources
Full Text Sources
