Microdeletion syndromes disclose replication timing alterations of genes unrelated to the missing DNA
- PMID: 19284877
- PMCID: PMC2660353
- DOI: 10.1186/1755-8166-2-11
Microdeletion syndromes disclose replication timing alterations of genes unrelated to the missing DNA
Abstract
Background: The temporal order of allelic replication is interrelated to the epigenomic profile. A significant epigenetic marker is the asynchronous replication of monoallelically-expressed genes versus the synchronous replication of biallelically-expressed genes. The present study sought to determine whether a microdeletion in the genome affects epigenetic profiles of genes unrelated to the missing segment. In order to test this hypothesis, we checked the replication patterns of two genes - SNRPN, a normally monoallelically expressed gene (assigned to 15q11.13), and the RB1, an archetypic biallelically expressed gene (assigned to 13.q14) in the genomes of patients carrying the 22q11.2 deletion (DiGeorge/Velocardiofacial syndrome) and those carrying the 7q11.23 deletion (Williams syndrome).
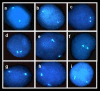
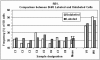
Results: The allelic replication timing was determined by fluorescence in situ hybridization (FISH) technology performed on peripheral blood cells. As expected, in the cells of normal subjects the frequency of cells showing asynchronous replication for SNRPN was significantly (P < 10-12) higher than the corresponding value for RB1. In contrast, cells of the deletion-carrying patients exhibited a reversal in this replication pattern: there was a significantly lower frequency of cells engaging in asynchronous replication for SNRPN than for RB1 (P < 10-4 and P < 10-3 for DiGeorge/Velocardiofacial and Williams syndromes, respectively). Accordingly, the significantly lower frequency of cells showing asynchronous replication for SNRPN than for RB1 is a new epigenetic marker distinguishing these deletion syndrome genotypes from normal ones.
Conclusion: In cell samples of each deletion-carrying individual, an aberrant, reversed pattern of replication is delineated, namely, where a monoallelic gene replicates more synchronously than a biallelic gene. This inverted pattern, which appears to be non-deletion-specific, clearly distinguishes cells of deletion-carriers from normal ones. As such, it offers a potential epigenetic marker for suspecting a hidden microdeletion that is too small to be detected by conventional karyotyping methods.
Figures




Similar articles
-
Aberrant allele-specific replication, independent of parental origin, in blood cells of cancer patients.BMC Cancer. 2008 Dec 25;8:390. doi: 10.1186/1471-2407-8-390. BMC Cancer. 2008. PMID: 19109880 Free PMC article.
-
Replication timing regulation in adults with chromosomal balance rearrangements.Genet Mol Res. 2015 Jul 14;14(3):7833-40. doi: 10.4238/2015.July.14.9. Genet Mol Res. 2015. PMID: 26214464
-
Modified allelic replication in lymphocytes of patients with neurofibromatosis type 1.Cancer Genet Cytogenet. 2003 Jun;143(2):133-9. doi: 10.1016/s0165-4608(02)00858-0. Cancer Genet Cytogenet. 2003. PMID: 12781447
-
22q11.2 deletion syndrome: DiGeorge, velocardiofacial, and conotruncal anomaly face syndromes.Curr Opin Pediatr. 2001 Oct;13(5):465-72. doi: 10.1097/00008480-200110000-00014. Curr Opin Pediatr. 2001. PMID: 11801894 Review.
-
A deletion and a duplication in distal 22q11.2 deletion syndrome region. Clinical implications and review.BMC Med Genet. 2009 Jun 2;10:48. doi: 10.1186/1471-2350-10-48. BMC Med Genet. 2009. PMID: 19490635 Free PMC article. Review.
Cited by
-
X-chromosome terminal deletion in a female with premature ovarian failure: Haploinsufficiency of X-linked genes as a possible explanation.Mol Cytogenet. 2010 Jul 20;3:14. doi: 10.1186/1755-8166-3-14. Mol Cytogenet. 2010. PMID: 20646274 Free PMC article.
-
Ontogenetic variation of the human genome.Curr Genomics. 2010 Sep;11(6):420-5. doi: 10.2174/138920210793175958. Curr Genomics. 2010. PMID: 21358986 Free PMC article.
-
Comprehensive analysis of DNA replication timing across 184 cell lines suggests a role for MCM10 in replication timing regulation.Hum Mol Genet. 2022 Aug 25;31(17):2899-2917. doi: 10.1093/hmg/ddac082. Hum Mol Genet. 2022. PMID: 35394024 Free PMC article.
References
-
- Shaffer LG, Ledbetter DH, Lupski JR. Molecular cytogenetics of contiguous gene syndromes: mechanisms and consequences of gene dosage imbalance. In: Scriver CR, Beaudet AL, Sly WS, Valle D, Childs B, Kinzler KW, Vogelstein B, editor. Metabolic and Molecular Basis of Inherited Disease. New York: McGraw Hill; 2001. pp. 1291–1324.
-
- Zahir F, Friedman JM. The impact of array genomic hybridization on mental retardation research: a review of current technologies and their clinical utility. Clin Genet. 2007;72:271–287. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

