Evolution of cell motility in an individual-based model of tumour growth
- PMID: 19285513
- PMCID: PMC2706369
- DOI: 10.1016/j.jtbi.2009.03.005
Evolution of cell motility in an individual-based model of tumour growth
Abstract
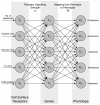
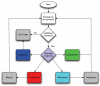
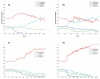
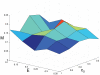
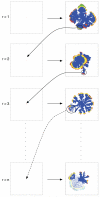
Tumour invasion is driven by proliferation and importantly migration into the surrounding tissue. Cancer cell motility is also critical in the formation of metastases and is therefore a fundamental issue in cancer research. In this paper we investigate the emergence of cancer cell motility in an evolving tumour population using an individual-based modelling approach. In this model of tumour growth each cell is equipped with a micro-environment response network that determines the behaviour or phenotype of the cell based on the local environment. The response network is modelled using a feed-forward neural network, which is subject to mutations when the cells divide. With this model we have investigated the impact of the micro-environment on the emergence of a motile invasive phenotype. The results show that when a motile phenotype emerges the dynamics of the model are radically changed and we observe faster growing tumours exhibiting diffuse morphologies. Further we observe that the emergence of a motile subclone can occur in a wide range of micro-environmental growth conditions. Iterated simulations showed that in identical growth conditions the evolutionary dynamics either converge to a proliferating or migratory phenotype, which suggests that the introduction of cell motility into the model changes the shape of fitness landscape on which the cancer cell population evolves and that it now contains several local maxima. This could have important implications for cancer treatments which focus on the gene level, as our results show that several distinct genotypes and critically distinct phenotypes can emerge and become dominant in the same micro-environment.
Figures



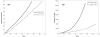

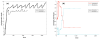

Similar articles
-
Bridging scales in cancer progression: mapping genotype to phenotype using neural networks.Semin Cancer Biol. 2015 Feb;30:30-41. doi: 10.1016/j.semcancer.2014.04.013. Epub 2014 May 12. Semin Cancer Biol. 2015. PMID: 24830623 Free PMC article. Review.
-
A hybrid cellular automaton model of clonal evolution in cancer: the emergence of the glycolytic phenotype.J Theor Biol. 2008 Feb 21;250(4):705-22. doi: 10.1016/j.jtbi.2007.10.038. Epub 2007 Nov 4. J Theor Biol. 2008. PMID: 18068192 Free PMC article.
-
Modelling evolutionary cell behaviour using neural networks: application to tumour growth.Biosystems. 2009 Feb;95(2):166-74. doi: 10.1016/j.biosystems.2008.10.007. Epub 2008 Nov 5. Biosystems. 2009. PMID: 19026711 Free PMC article.
-
An evolutionary hybrid cellular automaton model of solid tumour growth.J Theor Biol. 2007 Jun 21;246(4):583-603. doi: 10.1016/j.jtbi.2007.01.027. Epub 2007 Feb 12. J Theor Biol. 2007. PMID: 17374383 Free PMC article.
-
The matrix environmental and cell mechanical properties regulate cell migration and contribute to the invasive phenotype of cancer cells.Rep Prog Phys. 2019 Jun;82(6):064602. doi: 10.1088/1361-6633/ab1628. Epub 2019 Apr 4. Rep Prog Phys. 2019. PMID: 30947151 Review.
Cited by
-
Bridging scales in cancer progression: mapping genotype to phenotype using neural networks.Semin Cancer Biol. 2015 Feb;30:30-41. doi: 10.1016/j.semcancer.2014.04.013. Epub 2014 May 12. Semin Cancer Biol. 2015. PMID: 24830623 Free PMC article. Review.
-
Tumor evolution in space: the effects of competition colonization tradeoffs on tumor invasion dynamics.Front Oncol. 2013 Mar 6;3:45. doi: 10.3389/fonc.2013.00045. eCollection 2013. Front Oncol. 2013. PMID: 23508890 Free PMC article.
-
PhysiBoSS: a multi-scale agent-based modelling framework integrating physical dimension and cell signalling.Bioinformatics. 2019 Apr 1;35(7):1188-1196. doi: 10.1093/bioinformatics/bty766. Bioinformatics. 2019. PMID: 30169736 Free PMC article.
-
Cross-scale, cross-pathway evaluation using an agent-based non-small cell lung cancer model.Bioinformatics. 2009 Sep 15;25(18):2389-96. doi: 10.1093/bioinformatics/btp416. Epub 2009 Jul 4. Bioinformatics. 2009. PMID: 19578172 Free PMC article.
-
Using DNA methylation patterns to infer tumor ancestry.PLoS One. 2010 Aug 9;5(8):e12002. doi: 10.1371/journal.pone.0012002. PLoS One. 2010. PMID: 20711251 Free PMC article.
References
-
- Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. The Cell. 3rd Edition. Garland Publishing; New York: 1994. pp. 1173–1175. Ch. 22: Differentiated Cells and the Maintenance of Tissue.
-
- Anderson A, B.D S, Young I, Griffiths B. Nematode movement along a chemical gradient in a structurally heterogeneous environment. 2. theory. Fundamental and applied nematology. 1997;20:165–172.
-
- Anderson ARA. A hybrid mathematical model of solid tumour invasion: the importance of cell adhesion. Math. Med. Biol. 2005;22:163–186. - PubMed
-
- Anderson ARA, Chaplain M. Continuous and discrete mathematical models of tumor-induced angiogenesis. Bull. Math. Bio. 1998;6:857–899. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

