A functional genomic screen identifies cellular cofactors of hepatitis C virus replication
- PMID: 19286138
- PMCID: PMC2756022
- DOI: 10.1016/j.chom.2009.02.001
A functional genomic screen identifies cellular cofactors of hepatitis C virus replication
Abstract
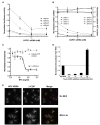
Hepatitis C virus (HCV) chronically infects 3% of the world's population, and complications from HCV are the leading indication for liver transplantation. Given the need for better anti-HCV therapies, one strategy is to identify and target cellular cofactors of the virus lifecycle. Using a genome-wide siRNA library, we identified 96 human genes that support HCV replication, with a significant number of them being involved in vesicle organization and biogenesis. Phosphatidylinositol 4-kinase PI4KA and multiple subunits of the COPI vesicle coat complex were among the genes identified. Consistent with this, pharmacologic inhibitors of COPI and PI4KA blocked HCV replication. Targeting hepcidin, a peptide critical for iron homeostasis, also affected HCV replication, which may explain the known dysregulation of iron homeostasis in HCV infection. The host cofactors for HCV replication identified in this study should serve as a useful resource in delineating new targets for anti-HCV therapies.
Figures


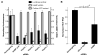
References
-
- Ahn J, Chung KS, Kim DU, Won M, Kim L, Kim KS, Nam M, Choi SJ, Kim HC, Yoon M, et al. Systematic identification of hepatocellular proteins interacting with NS5A of the hepatitis C virus. J Biochem Mol Biol. 2004;37:741–748. - PubMed
-
- Aoki CA, Rossaro L, Ramsamooj R, Brandhagen D, Burritt MF, Bowlus CL. Liver hepcidin mRNA correlates with iron stores, but not inflammation, in patients with chronic hepatitis C. J Clin Gastroenterol. 2005;39:71–74. - PubMed
-
- Balla A, Balla T. Phosphatidylinositol 4-kinases: old enzymes with emerging functions. Trends Cell Biol. 2006;16:351–361. - PubMed
-
- Balla T, Downing GJ, Jaffe H, Kim S, Zólyomi A, Catt KJ. Isolation and molecular cloning of wortmannin-sensitive bovine type III phosphatidylinositol 4-kinases. J Biol Chem. 1997;272:18358–18366. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

