A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock
- PMID: 19286557
- PMCID: PMC4259050
- DOI: 10.1126/science.1167206
A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock
Erratum in
- Science. 2009 Oct 16;326(5951):366
Abstract
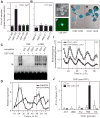
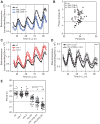
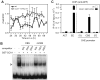
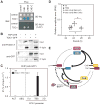
Transcriptional feedback loops constitute the molecular circuitry of the plant circadian clock. In Arabidopsis, a core loop is established between CCA1 and TOC1. Although CCA1 directly represses TOC1, the TOC1 protein has no DNA binding domains, which suggests that it cannot directly regulate CCA1. We established a functional genomic strategy that led to the identification of CHE, a TCP transcription factor that binds specifically to the CCA1 promoter. CHE is a clock component partially redundant with LHY in the repression of CCA1. The expression of CHE is regulated by CCA1, thus adding a CCA1/CHE feedback loop to the Arabidopsis circadian network. Because CHE and TOC1 interact, and CHE binds to the CCA1 promoter, a molecular linkage between TOC1 and CCA1 gene regulation is established.
Figures
Comment in
-
Circadian rhythms. Linking the loops.Science. 2009 Mar 13;323(5920):1440-1. doi: 10.1126/science.1171418. Science. 2009. PMID: 19286545 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

