Clathrin regulates the association of PIPKIgamma661 with the AP-2 adaptor beta2 appendage
- PMID: 19287005
- PMCID: PMC2679492
- DOI: 10.1074/jbc.M901017200
Clathrin regulates the association of PIPKIgamma661 with the AP-2 adaptor beta2 appendage
Abstract
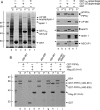
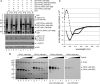
The AP-2 clathrin adaptor differs fundamentally from the related AP-1, AP-3, and AP-4 sorting complexes because membrane deposition does not depend directly on an Arf family GTPase. Instead phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P(2)) appears to act as the principal compartmental cue for AP-2 placement at the plasma membrane as well as for the docking of numerous other important clathrin coat components at the nascent bud site. This PtdIns(4,5)P(2) dependence makes type I phosphatidylinositol 4-phosphate 5-kinases (PIPKIs) lynchpin enzymes in the assembly of clathrin-coated structures at the cell surface. PIPKIgamma is the chief 5-kinase at nerve terminals, and here we show that the 26-amino acid, alternatively spliced C terminus of PIPKIgamma661 is an intrinsically unstructured polypeptide that binds directly to the sandwich subdomain of the AP-2 beta2 subunit appendage. An aromatic side chain-based, extended interaction motif that also includes the two bulky C-terminal residues of the short PIPKIgamma635 variant is necessary for beta2 appendage engagement. The clathrin heavy chain accesses the same contact surface on the AP-2 beta2 appendage, but because of additional clathrin binding sites located within the unstructured hinge segment of the beta2 subunit, clathrin binds the beta2 chain with a higher apparent affinity than PIPKIgamma661. A clathrin-regulated interaction with AP-2 could allow PIPKIgamma661 to be strategically positioned for regional PtdIns(4,5)P(2) generation during clathrin-coated vesicle assembly at the synapse.
Figures


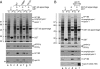




References
-
- Di Paolo, G., and De Camilli, P. (2006) Nature 443 651-657 - PubMed
-
- Maldonado-Baez, L., and Wendland, B. (2006) Trends Cell Biol. 16 505-513 - PubMed
-
- Schmid, E. M., and McMahon, H. T. (2007) Nature 448 883-888 - PubMed
-
- Ungewickell, E. J., and Hinrichsen, L. (2007) Curr. Opin. Cell Biol. 19 417-425 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

