Phenotypes, genotypes and disease susceptibility associated with gene copy number variations: complement C4 CNVs in European American healthy subjects and those with systemic lupus erythematosus
- PMID: 19287147
- PMCID: PMC2709077
- DOI: 10.1159/000184700
Phenotypes, genotypes and disease susceptibility associated with gene copy number variations: complement C4 CNVs in European American healthy subjects and those with systemic lupus erythematosus
Abstract
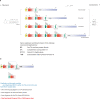
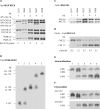
A new paradigm in human genetics is high frequencies of inter-individual variations in copy numbers of specific genomic DNA segments. Such common copy number variation (CNV) loci often contain genes engaged in host-environment interaction including those involved in immune effector functions. DNA sequences within a CNV locus often share a high degree of identity but beneficial or deleterious polymorphic variants are present among different individuals. Thus, common gene CNVs can contribute, both qualitatively and quantitatively, to a spectrum of phenotypic variants. In this review we describe the phenotypic and genotypic diversities of complement C4 created by copy number variations of RCCX modules (RP-C4-CYP21-TNX) and size dichotomy of C4 genes. A direct outcome of C4 CNV is the generation of two classes of polymorphic proteins, C4A and C4B, with differential chemical reactivities towards peptide or carbohydrate antigens, and a range of C4 plasma protein concentrations (from 15 to 70 mg/dl) among healthy subjects. Deliberate molecular genetic studies enabled development of definitive techniques to determine exact patterns of RCCX modular variations, copy numbers of long and short C4A and C4B genes by Southern blot analyses or by real-time quantitative PCR. It is found that in healthy European Americans, the total C4 gene copy number per diploid genome ranges from 2 to 6: 60.8% of people with four copies of C4 genes, 27.2% with less than four copies, and 12% with more than four copies. Such a distribution is skewed towards the low copy number side in patients with systemic lupus erythematosus (SLE), a prototypic autoimmune disease with complex etiology. In SLE, the frequency of individuals with less than four copies of C4 is significantly increased (42.2%), while the frequency of those with more than four copies is decreased (6%). This decrease in total C4 gene copy number in SLE is due to increases in homozygous and heterozygous deficiencies of C4A but not C4B. Therefore, it is concluded that lower copy number of C4 is a risk factor for and higher gene copy number of C4 is a protective factor against SLE disease susceptibility.
Copyright 2009 S. Karger AG, Basel.
Figures

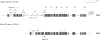

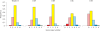
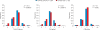
References
-
- Awdeh ZL, Raum D, Alper CA. Genetic polymorphism of human complement C4 and detection of heterozygotes. Nature. 1979;282:205–208. - PubMed
-
- Belt KT, Caroll MC, Porter RR. The structural basis of the multiple forms of human complement component C4. Cell. 1984;36:907–914. - PubMed
-
- Belt KT, Yu CY, Carroll MC, Porter RR. Polymorphism of human complement component C4. Immunogenetics. 1985;21:173–180. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

