Regulation of clock-controlled genes in mammals
- PMID: 19287494
- PMCID: PMC2654074
- DOI: 10.1371/journal.pone.0004882
Regulation of clock-controlled genes in mammals
Abstract
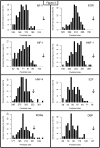
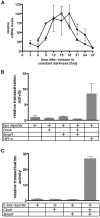
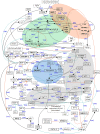
The complexity of tissue- and day time-specific regulation of thousands of clock-controlled genes (CCGs) suggests that many regulatory mechanisms contribute to the transcriptional output of the circadian clock. We aim to predict these mechanisms using a large scale promoter analysis of CCGs.Our study is based on a meta-analysis of DNA-array data from rodent tissues. We searched in the promoter regions of 2065 CCGs for highly overrepresented transcription factor binding sites. In order to compensate the relatively high GC-content of CCG promoters, a novel background model to avoid a bias towards GC-rich motifs was employed. We found that many of the transcription factors with overrepresented binding sites in CCG promoters exhibit themselves circadian rhythms. Among the predicted factors are known regulators such as CLOCKratioBMAL1, DBP, HLF, E4BP4, CREB, RORalpha and the recently described regulators HSF1, STAT3, SP1 and HNF-4alpha. As additional promising candidates of circadian transcriptional regulators PAX-4, C/EBP, EVI-1, IRF, E2F, AP-1, HIF-1 and NF-Y were identified. Moreover, GC-rich motifs (SP1, EGR, ZF5, AP-2, WT1, NRF-1) and AT-rich motifs (MEF-2, HMGIY, HNF-1, OCT-1) are significantly overrepresented in promoter regions of CCGs. Putative tissue-specific binding sites such as HNF-3 for liver, NKX2.5 for heart or Myogenin for skeletal muscle were found. The regulation of the erythropoietin (Epo) gene was analysed, which exhibits many binding sites for circadian regulators. We provide experimental evidence for its circadian regulated expression in the adult murine kidney. Basing on a comprehensive literature search we integrate our predictions into a regulatory network of core clock and clock-controlled genes. Our large scale analysis of the CCG promoters reveals the complexity and extensiveness of the circadian regulation in mammals. Results of this study point to connections of the circadian clock to other functional systems including metabolism, endocrine regulation and pharmacokinetics.
Conflict of interest statement
Figures

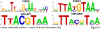
References
-
- Goldbeter A. Biochemical Oscillations and Cellular Rhythms: The Molecular Bases of Periodic and Chaotic Behaviour. Cambridge, United Kingdom: Cambridge University Press; 1996.
-
- Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, et al. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–82. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

