Thermodynamic pathways to genome spatial organization in the cell nucleus
- PMID: 19289043
- PMCID: PMC2717292
- DOI: 10.1016/j.bpj.2008.12.3919
Thermodynamic pathways to genome spatial organization in the cell nucleus
Abstract
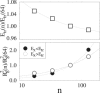
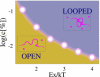
The architecture of the eukaryotic genome is characterized by a high degree of spatial organization. Chromosomes occupy preferred territories correlated to their state of activity and, yet, displace their genes to interact with remote sites in complex patterns requiring the orchestration of a huge number of DNA loci and molecular regulators. Far from random, this organization serves crucial functional purposes, but its governing principles remain elusive. By computer simulations of a statistical mechanics model, we show how architectural patterns spontaneously arise from the physical interaction between soluble binding molecules and chromosomes via collective thermodynamics mechanisms. Chromosomes colocalize, loops and territories form, and find their relative positions as stable thermodynamic states. These are selected by thermodynamic switches, which are regulated by concentrations/affinity of soluble mediators and by number/location of their attachment sites along chromosomes. Our thermodynamic switch model of nuclear architecture, thus, explains on quantitative grounds how well-known cell strategies of upregulation of DNA binding proteins or modification of chromatin structure can dynamically shape the organization of the nucleus.
Figures



References
-
- Cremer T., Cremer M., Dietzel S., Muller S., Solovei I. Chromosome territories—a functional nuclear landscape. Curr. Opin. Cell Biol. 2006;18:307–316. - PubMed
-
- Lanctôt C., Cheutin T., Cremer M., Cavalli G., Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat. Rev. Genet. 2007;8:104–115. - PubMed
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Meaburn K., Misteli T. Chromosome territories. Nature. 2007;445:379–381. - PubMed
-
- Fraser P., Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447:413–417. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

