Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy
- PMID: 19289046
- PMCID: PMC2717305
- DOI: 10.1016/j.bpj.2008.11.063
Structure and membrane interactions of the antibiotic peptide dermadistinctin K by multidimensional solution and oriented 15N and 31P solid-state NMR spectroscopy
Abstract
DD K, a peptide first isolated from the skin secretion of the Phyllomedusa distincta frog, has been prepared by solid-phase chemical peptide synthesis and its conformation was studied in trifluoroethanol/water as well as in the presence of sodium dodecyl sulfate and dodecylphosphocholine micelles or small unilamellar vesicles. Multidimensional solution NMR spectroscopy indicates an alpha-helical conformation in membrane environments starting at residue 7 and extending to the C-terminal carboxyamide. Furthermore, DD K has been labeled with (15)N at a single alanine position that is located within the helical core region of the sequence. When reconstituted into oriented phosphatidylcholine membranes the resulting (15)N solid-state NMR spectrum shows a well-defined helix alignment parallel to the membrane surface in excellent agreement with the amphipathic character of DD K. Proton-decoupled (31)P solid-state NMR spectroscopy indicates that the peptide creates a high level of disorder at the level of the phospholipid headgroup suggesting that DD K partitions into the bilayer where it severely disrupts membrane packing.
Figures
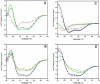


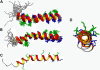
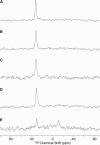
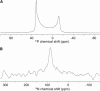

References
-
- Zasloff M. Antimicrobial peptides of multicellular organisms. Nature. 2002;415:389–395. - PubMed
-
- Boman H.G. Antibacterial peptides: basic facts and emerging concepts. J. Intern. Med. 2003;254:197–215. - PubMed
-
- Powers J.P., Hancock R.E. The relationship between peptide structure and antibacterial activity. Peptides. 2003;24:1681–1691. - PubMed
-
- Lehrer R.I. Primate defensins. Nat. Rev. Microbiol. 2004;2:727–738. - PubMed
-
- Garcia-Olmedo F., Molina A., Alamillo J.M., Rodriguez-Palenzuela P. Plant defense peptides. Biopolymers. 1999;47:479–491. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

