A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation
- PMID: 19289494
- PMCID: PMC2681996
- DOI: 10.1128/MCB.00042-09
A mouse PRMT1 null allele defines an essential role for arginine methylation in genome maintenance and cell proliferation
Erratum in
-
Correction for Yu et al., "A Mouse PRMT1 Null Allele Defines an Essential Role for Arginine Methylation in Genome Maintenance and Cell Proliferation".Mol Cell Biol. 2017 Aug 11;37(17):e00298-17. doi: 10.1128/MCB.00298-17. Print 2017 Sep 1. Mol Cell Biol. 2017. PMID: 28801457 Free PMC article. No abstract available.
Abstract
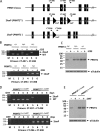
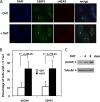
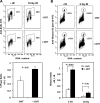
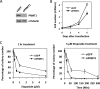
Protein arginine methyltransferase 1 (PRMT1) is the major enzyme that generates monomethylarginine and asymmetrical dimethylarginine. We report here a conditional null allele of PRMT1 in mice and that the loss of PRMT1 expression leads to embryonic lethality. Using the Cre/lox-conditional system, we show that the loss of PRMT1 in mouse embryonic fibroblasts (MEFs) leads to the loss of arginine methylation of substrates harboring a glycine-arginine rich motif, including Sam68 and MRE11. The loss of PRMT1 in MEFs leads to spontaneous DNA damage, cell cycle progression delay, checkpoint defects, aneuploidy, and polyploidy. We show using a 4-hydroxytamoxifen-inducible Cre that the loss of PRMT1 in MEFs leads to a higher incidence of chromosome losses, gains, structural rearrangements, and polyploidy, as documented by spectral karyotyping. Using PRMT1 small interfering RNA in U2OS cells, we further show that PRMT1-deficient cells are hypersensitive to the DNA damaging agent etoposide and exhibit a defect in the recruitment of the homologous recombination RAD51 recombinase to DNA damage foci. Taken together, these data show that PRMT1 is required for genome integrity and cell proliferation. Our findings also suggest that arginine methylation by PRMT1 is a key posttranslational modification in the DNA damage response pathway in proliferating mammalian cells.
Figures





References
-
- Adams, M. M., B. Wang, Z. Xia, J. C. Morales, X. Lu, L. A. Donehower, D. A. Bochar, S. J. Elledge, and P. B. Carpenter. 2005. 53BP1 oligomerization is independent of its methylation by PRMT1. Cell Cycle 41854-1861. - PubMed
-
- An, W., J. Kim, and R. G. Roeder. 2004. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell 117735-748. - PubMed
-
- Baumann, P., and S. West. 1998. Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem. Sci. 23247-251. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
