Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals
- PMID: 19289835
- PMCID: PMC2664036
- DOI: 10.1073/pnas.0810909106
Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals
Abstract
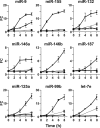
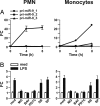
Inflammation involves a coordinated, sequential, and self limiting sequence of events controlled by positive and negative regulatory mechanisms. Recent studies have shown that microRNAs (miRNAs), an evolutionarily conserved class of endogenous 22-nucleotide noncoding RNAs, contribute to the regulation of inflammation by repressing gene expression at the posttranscriptional level. In this study, we characterize the profile of miRNAs induced by LPS in human polymorphonuclear neutrophils (PMN) and monocytes. In particular, we identify miR-9 as the only miRNA (among 365 analyzed) up-regulated in both cell types after TLR4 activation. miR-9 is also induced by TLR2 and TLR7/8 agonists and by the proinflammatory cytokines TNF-alpha and IL-1beta, but not by IFNgamma. Among the 3 different genes encoding miR-9 precursors in humans, we show that LPS selectively induces the transcription of miR-9-1 located in the CROC4 locus, in a MyD88- and NF-kappaB-dependent manner. In PMN and monocytes, LPS regulates NFKB1 at both the transcriptional and posttranscriptional levels, and a conserved miR-9 seed sustained a miR-9-dependent inhibition of the NFKB1 transcript. Overall, these data suggest that TLR4-activated NF-kappaB rapidly increases the expression of miR-9 that operates a feedback control of the NF-kappaB-dependent responses by fine tuning the expression of a key member of the NF-kappaB family.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. - PubMed
-
- Akira S, Takeda K. Toll-like receptor signalling. Nat Rev Immunol. 2004;4:499–511. - PubMed
-
- O'Neill LA. How Toll-like receptors signal: What we know and what we don't know. Curr Opin Immunol. 2006;18:3–9. - PubMed
-
- Liew FY, Xu D, Brint EK, O'Neill LA. Negative regulation of toll-like receptor-mediated immune responses. Nat Rev Immunol. 2005;5:446–458. - PubMed
-
- Mantovani A, Locati M, Polentarutti N, Vecchi A, Garlanda C. Extracellular and intracellular decoys in the tuning of inflammatory cytokines and Toll-like receptors: The new entry TIR8/SIGIRR. J Leukoc Biol. 2004;75:738–742. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

