MicroRNA expression profiles predictive of human renal allograft status
- PMID: 19289845
- PMCID: PMC2663998
- DOI: 10.1073/pnas.0813121106
MicroRNA expression profiles predictive of human renal allograft status
Abstract
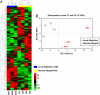
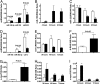
Immune rejection of organ transplants is a life-threatening complication and is exemplified by alterations in the expression of protein-encoding genes. Because microRNAs (miRNAs) regulate the expression of genes implicated in adaptive immunity, we investigated whether acute rejection (AR) is associated with alterations in miRNA expression within allografts and whether expression profiles are diagnostic of AR and predict allograft function. Seven of 33 renal allograft biopsies (12 AR and 21 normal) were profiled using microfluidic cards containing 365 mature human miRNAs (training set), and a subset of differentially expressed miRNAs were quantified in the remaining 26 allograft biopsies (validation set). We found a strong association between intragraft expression of miRNAs and messenger RNAs (mRNAs), and that AR, and renal allograft function, could be predicted with a high level of precision using intragraft levels of miRNAs. Our investigation of miRNA expression in normal human peripheral blood mononuclear cells (PBMCs) showed that miRNAs (miR-142-5p, -155, and -223) overexpressed in AR biopsies are highly expressed in PBMCs, and that stimulation with the mitogen phytohaemagglutinin results in an increase in the abundance of miR-155 and a decrease in miR-223 and let-7c. Quantification of miRNAs in primary cultures of human renal epithelial cells (HRECs) showed that miR-30a-3p, -10b, and let-7c are highly expressed in HRECs, and that stimulation results in a decreased expression of miR-30a-3p. Our studies, in addition to suggesting a cellular basis for the altered intragraft expression of miRNAs, propose that miRNA expression patterns may serve as biomarkers of human renal allograft status.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


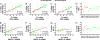

References
-
- Grosshans H, Filipowicz W. Molecular biology: The expanding world of small RNAs. Nature. 2008;451:414–416. - PubMed
-
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. - PubMed
-
- Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

