Cohesins: chromatin architects in chromosome segregation, control of gene expression and much more
- PMID: 19290475
- PMCID: PMC11115881
- DOI: 10.1007/s00018-009-0004-8
Cohesins: chromatin architects in chromosome segregation, control of gene expression and much more
Abstract
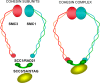
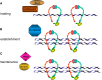
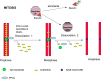
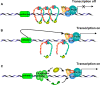
Cells have evolved to develop molecules and control mechanisms that guarantee correct chromosome segregation and ensure the proper distribution of genetic material to daughter cells. In this sense, the establishment, maintenance, and removal of sister chromatid cohesion is one of the most fascinating and dangerous processes in the life of a cell because errors in the control of these processes frequently lead to cell death or aneuploidy. The main protagonist in this mechanism is a four-protein complex denominated the cohesin complex. In the last 10 years, we have improved our understanding of the key players in the regulation of sister chromatid cohesion during cell division in mitosis and meiosis. The last 2 years have seen an increase in evidence showing that cohesins have important functions in non-dividing cells, revealing new, unexplored roles for these proteins in the control of gene expression, development, and other essential cell functions in mammals.
Figures

Similar articles
-
Sister chromatid cohesion control and aneuploidy.Cytogenet Genome Res. 2011;133(2-4):223-33. doi: 10.1159/000323507. Epub 2011 Jan 19. Cytogenet Genome Res. 2011. PMID: 21252490 Review.
-
[Sister chromatid cohesion complex in Eukaryota].Postepy Biochem. 2010;56(1):41-54. Postepy Biochem. 2010. PMID: 20499680 Review. Polish.
-
Centromeric Cohesin: Molecular Glue and Much More.Prog Mol Subcell Biol. 2017;56:485-513. doi: 10.1007/978-3-319-58592-5_20. Prog Mol Subcell Biol. 2017. PMID: 28840250 Review.
-
Cohesin complexes and sister chromatid cohesion in mammalian meiosis.Genome Dyn. 2009;5:94-116. doi: 10.1159/000166622. Genome Dyn. 2009. PMID: 18948710 Review.
-
The sister bonding of duplicated chromosomes.Semin Cell Dev Biol. 2011 Aug;22(6):566-71. doi: 10.1016/j.semcdb.2011.03.013. Epub 2011 Apr 7. Semin Cell Dev Biol. 2011. PMID: 21497666 Free PMC article. Review.
Cited by
-
Dynamics of cohesin subunits in grasshopper meiotic divisions.Chromosoma. 2013 Mar;122(1-2):77-91. doi: 10.1007/s00412-012-0393-6. Epub 2013 Jan 4. Chromosoma. 2013. PMID: 23283389
-
Involvement of the Cohesin Cofactor PDS5 (SPO76) During Meiosis and DNA Repair in Arabidopsis thaliana.Front Plant Sci. 2015 Dec 1;6:1034. doi: 10.3389/fpls.2015.01034. eCollection 2015. Front Plant Sci. 2015. PMID: 26648949 Free PMC article.
-
Identification of heat shock factor 1 molecular and cellular targets during embryonic and adult female meiosis.Mol Cell Biol. 2011 Aug;31(16):3410-23. doi: 10.1128/MCB.05237-11. Epub 2011 Jun 20. Mol Cell Biol. 2011. PMID: 21690297 Free PMC article.
-
Nonallelic transcriptional roles of CTCF and cohesins at imprinted loci.Mol Cell Biol. 2011 Aug;31(15):3094-104. doi: 10.1128/MCB.01449-10. Epub 2011 May 31. Mol Cell Biol. 2011. PMID: 21628529 Free PMC article.
-
The spectrum of genetic mutations in myelodysplastic syndrome: Should we update prognostication?EJHaem. 2021 Nov 1;3(1):301-313. doi: 10.1002/jha2.317. eCollection 2022 Feb. EJHaem. 2021. PMID: 35846202 Free PMC article. Review.
References
-
- Carramolino L, Lee BC, Zaballos A, Peled A, Barthelemy I, Shav-Tal Y, Prieto I, Carmi P, Gothelf Y, Gonzalez de Buitrago G, Aracil M, Marquez G, Barbero JL, Zipori D. SA-1, a nuclear protein encoded by one member of a novel gene family: molecular cloning and detection in hemopoietic organs. Gene. 1997;195:151–159. doi: 10.1016/S0378-1119(97)00121-2. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

