MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine
- PMID: 19290790
- PMCID: PMC2705205
- DOI: 10.2217/14622416.10.3.399
MicroRNA polymorphisms: the future of pharmacogenomics, molecular epidemiology and individualized medicine
Abstract
Referred to as the micromanagers of gene expression, microRNAs (miRNAs) are evolutionarily conserved small noncoding RNAs. Polymorphisms in the miRNA pathway (miR-polymorphisms) are emerging as powerful tools to study the biology of a disease and have the potential to be used in disease prognosis and diagnosis. Detection of miR-polymorphisms holds promise in the field of miRNA pharmacogenomics, molecular epidemiology and for individualized medicine. MiRNA pharmacogenomics can be defined as the study of miRNAs and polymorphisms affecting miRNA function in order to predict drug behavior and to improve drug efficacy. Advancements in the miRNA field indicate the clear involvement of miRNAs and genetic variations within the miRNA pathway in the progression and prognosis of diseases such as cancer, neurological disorders, muscular hypertrophy, gastric mucosal atrophy, cardiovascular disease and Type II diabetes. Various algorithms are available to predict miRNA-target mRNA sites; however, it is advisable to use multiple algorithms to confirm the predictions. Polymorphisms that may potentially affect miRNA-mediated regulation of the cell can be present not only in the 3 -UTR of a miRNA target gene, but also in the genes involved in miRNA biogenesis and in pri-, pre- and mature-miRNA sequences. A polymorphism in processed miRNAs may affect expression of several genes and have serious consequences, whereas a polymorphism in miRNA target site, in the 3 -UTR of the target mRNA, may be more target and/or pathway specific. In this review, we for the first time suggest a classification of miRNA polymorphisms/mutations. We also describe the importance and implications of miR-polymorphisms in gene regulation, disease progression, pharmacogenomics and molecular epidemiology.
Figures



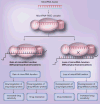
References
-
- Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. Landmark paper that identified small noncoding microRNA (miRNA) genes. - PubMed
-
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. Landmark paper that identified small noncoding miRNA genes. - PubMed
-
- Lee RC, Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. Landmark paper that identified small noncoding miRNA genes. - PubMed
-
- Mishra PJ, Humeniuk R, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci USA. 2007;104:13513–13518. Established the role of a miRNA polymorphism in drug-resistance; the term miRSNP was coined and defined. MiR-24 destabilizes the target dihydrofolate reductase (DHFR) mRNA. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
