Carbohydrate recognition by boronolectins, small molecules, and lectins
- PMID: 19291708
- PMCID: PMC2829346
- DOI: 10.1002/med.20155
Carbohydrate recognition by boronolectins, small molecules, and lectins
Abstract
Carbohydrates are known to mediate a large number of biological and pathological events. Small and macromolecules capable of carbohydrate recognition have great potentials as research tools, diagnostics, vectors for targeted delivery of therapeutic and imaging agents, and therapeutic agents. However, this potential is far from being realized. One key issue is the difficulty in the development of "binders" capable of specific recognition of carbohydrates of biological relevance. This review discusses systematically the general approaches that are available in developing carbohydrate sensors and "binders/receptors," and their applications. The focus is on discoveries during the last 5 years.
(c) 2009 Wiley Periodicals, Inc.
Figures
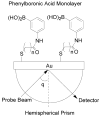








Similar articles
-
Fluorescent indolylboronic acids that are useful reporters for the synthesis of boronolectins.Chem Biol Drug Des. 2006 Feb;67(2):137-44. doi: 10.1111/j.1747-0285.2005.00338.x. Chem Biol Drug Des. 2006. PMID: 16492161
-
Boronolectins and fluorescent boronolectins: an examination of the detailed chemistry issues important for the design.Med Res Rev. 2005 Sep;25(5):490-520. doi: 10.1002/med.20038. Med Res Rev. 2005. PMID: 16025498 Review.
-
High-affinity carbohydrate binding by trimeric benzoboroxoles measured on carbohydrate arrays.Chembiochem. 2014 Nov 3;15(16):2450-7. doi: 10.1002/cbic.201402297. Epub 2014 Sep 10. Chembiochem. 2014. PMID: 25209734
-
Fluorescent carbohydrate probes for cell lectins.Spectrochim Acta A Mol Biomol Spectrosc. 2001 Sep 14;57(11):2285-96. doi: 10.1016/s1386-1425(01)00478-4. Spectrochim Acta A Mol Biomol Spectrosc. 2001. PMID: 11603844
-
Fluorescently labelled glycans and their applications.Glycoconj J. 2015 Nov;32(8):559-74. doi: 10.1007/s10719-015-9611-9. Epub 2015 Aug 4. Glycoconj J. 2015. PMID: 26239924 Review.
Cited by
-
Boronic acid porphyrin receptor for ginsenoside sensing.Org Lett. 2010 Nov 5;12(21):4804-7. doi: 10.1021/ol1019647. Org Lett. 2010. PMID: 20860384 Free PMC article.
-
In situ recognition of cell-surface glycans and targeted imaging of cancer cells.Sci Rep. 2013;3:2679. doi: 10.1038/srep02679. Sci Rep. 2013. PMID: 24042097 Free PMC article.
-
Nanoscale controlled architecture for development of ultrasensitive lectin biosensors applicable in glycomics.Anal Methods. 2014 Jul 21;6(14):4922-4931. doi: 10.1039/C4AY00495G. Epub 2014 Jun 11. Anal Methods. 2014. PMID: 27231486 Free PMC article.
-
Affinity enhancement by dendritic side chains in synthetic carbohydrate receptors.Angew Chem Int Ed Engl. 2015 Feb 9;54(7):2057-61. doi: 10.1002/anie.201409124. Epub 2015 Jan 21. Angew Chem Int Ed Engl. 2015. PMID: 25645064 Free PMC article.
-
Graphene Hybrid Materials for Controlling Cellular Microenvironments.Materials (Basel). 2020 Sep 10;13(18):4008. doi: 10.3390/ma13184008. Materials (Basel). 2020. PMID: 32927729 Free PMC article. Review.
References
-
- Alavi A, Axford JS, editors. Glycoimmunology. Vol. 376. New York: Plenum Press; 1995.
-
- Timmer MS, Stocker BL, Seeberger PH. Probing Glycomics. Curr Opin Chem Biol. 2007;11(1):59–65. - PubMed
-
- Alvarez-Manilla G, Warren NL, Abney T, Atwood J, 3rd, Azadi P, York WS, Pierce M, Orlando R. Tools for Glycomics: Relative Quantitation of Glycans by Isotopic Permethylation using 13CH3I. Glycobiology. 2007;17(7):677–687. - PubMed
-
- Fukuda M. Cell Surface Carbohydrates: Cell-type Specific Expression. In: Fukuda M, Hindsgaul O, editors. Molecular Glycobiology . New York: Oxford University Press; 1994. pp. 1–52.
-
- Dennis JW. Changes in Glycosylation Associated with Malignant Transformation and Tumor Progression. In: Fukuda M, editor. Cell Surface Carbohydrates and Cell Development . Boca Raton: CRC Press; 1992. pp. 161–194.
Publication types
MeSH terms
Substances
Grants and funding
- DK55062/DK/NIDDK NIH HHS/United States
- CA113917/CA/NCI NIH HHS/United States
- CA122536/CA/NCI NIH HHS/United States
- GM084933/GM/NIGMS NIH HHS/United States
- R42 GM086925/GM/NIGMS NIH HHS/United States
- R01 GM084933/GM/NIGMS NIH HHS/United States
- CA88343/CA/NCI NIH HHS/United States
- N01-CO-27184/CO/NCI NIH HHS/United States
- R33 CA088343/CA/NCI NIH HHS/United States
- R21 CA088343/CA/NCI NIH HHS/United States
- R21 CA113917/CA/NCI NIH HHS/United States
- GM086925/GM/NIGMS NIH HHS/United States
- R21 CA123329/CA/NCI NIH HHS/United States
- CA123329/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

