The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation
- PMID: 19293276
- PMCID: PMC2685098
- DOI: 10.1093/nar/gkp166
The solution structure of the first PHD finger of autoimmune regulator in complex with non-modified histone H3 tail reveals the antagonistic role of H3R2 methylation
Abstract
Plant homeodomain (PHD) fingers are often present in chromatin-binding proteins and have been shown to bind histone H3 N-terminal tails. Mutations in the autoimmune regulator (AIRE) protein, which harbours two PHD fingers, cause a rare monogenic disease, autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (APECED). AIRE activates the expression of tissue-specific antigens by directly binding through its first PHD finger (AIRE-PHD1) to histone H3 tails non-methylated at K4 (H3K4me0). Here, we present the solution structure of AIRE-PHD1 in complex with H3K4me0 peptide and show that AIRE-PHD1 is a highly specialized non-modified histone H3 tail reader, as post-translational modifications of the first 10 histone H3 residues reduce binding affinity. In particular, H3R2 dimethylation abrogates AIRE-PHD1 binding in vitro and reduces the in vivo activation of AIRE target genes in HEK293 cells. The observed antagonism by R2 methylation on AIRE-PHD1 binding is unique among the H3K4me0 histone readers and represents the first case of epigenetic negative cross-talk between non-methylated H3K4 and methylated H3R2. Collectively, our results point to a very specific histone code responsible for non-modified H3 tail recognition by AIRE-PHD1 and describe at atomic level one crucial step in the molecular mechanism responsible for antigen expression in the thymus.
Figures
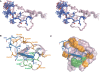
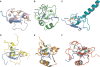

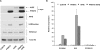
References
-
- Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem. Sci. 2006;31:35–40. - PubMed
-
- Aasland R, Gibson TJ, Stewart AF. The PHD finger: implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995;20:56–59. - PubMed
-
- Zhang Y. It takes a PHD to interpret histone methylation. Nat. Struct. Mol. Biol. 2006;13:572–574. - PubMed
-
- Mellor J. It takes a PHD to read the histone code. Cell. 2006;126:22–24. - PubMed
-
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

