Enterohemorrhagic Escherichia coli O157:H7 gene expression profiling in response to growth in the presence of host epithelia
- PMID: 19293938
- PMCID: PMC2654852
- DOI: 10.1371/journal.pone.0004889
Enterohemorrhagic Escherichia coli O157:H7 gene expression profiling in response to growth in the presence of host epithelia
Abstract
Background: The pathogenesis of enterohemorrhagic Escherichia coli (EHEC) O157:H7 infection is attributed to virulence factors encoded on multiple pathogenicity islands. Previous studies have shown that EHEC O157:H7 modulates host cell signal transduction cascades, independent of toxins and rearrangement of the cytoskeleton. However, the virulence factors and mechanisms responsible for EHEC-mediated subversion of signal transduction remain to be determined. Therefore, the purpose of this study was to first identify differentially regulated genes in response to EHEC O157:H7 grown in the presence of epithelial cells, compared to growth in the absence of epithelial cells (that is, growth in minimal essential tissue culture medium alone, minimal essential tissue culture medium in the presence of 5% CO(2), and Penassay broth alone) and, second, to identify EHEC virulence factors responsible for pathogen modulation of host cell signal transduction.
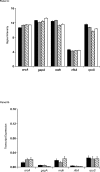
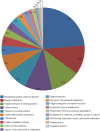
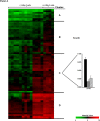
Methodology/principal findings: Overnight cultures of EHEC O157:H7 were incubated for 6 hr at 37 degrees C in the presence or absence of confluent epithelial (HEp-2) cells. Total RNA was then extracted and used for microarray analyses (Affymetrix E. coli Genome 2.0 gene chips). Relative to bacteria grown in each of the other conditions, EHEC O157:H7 cultured in the presence of cultured epithelial cells displayed a distinct gene-expression profile. A 2.0-fold increase in the expression of 71 genes and a 2.0-fold decrease in expression of 60 other genes were identified in EHEC O157:H7 grown in the presence of epithelial cells, compared to bacteria grown in media alone.
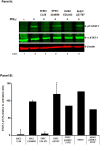
Conclusion/significance: Microarray analyses and gene deletion identified a protease on O-island 50, gene Z1787, as a potential virulence factor responsible for mediating EHEC inhibition of the interferon (IFN)-gamma-Jak1,2-STAT-1 signal transduction cascade. Up-regulated genes provide novel targets for use in developing strategies to interrupt the infectious process.
Conflict of interest statement
Figures


References
-
- Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. - PubMed
-
- Serna AT, Boedeker EC. Pathogenesis and treatment of Shiga toxin-producing Escherichia coli infections. Curr Opin Gastroenterol. 2008;24:38–47. - PubMed
-
- Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. - PubMed
-
- Jandu N, Shen S, Wickham ME, Prajapati R, Finlay BB, et al. Multiple seropathotypes of verotoxin-producing Escherichia coli (VTEC) disrupt interferon gamma-induced tyrosine phosphorylation of signal transducer and activator of transcription (Stat)-1. Microb Pathog. 2007;42:62–71. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

